

 |
 |
Perception and Signal Transduction of Rhizobial Nod Factors |
||||||||||||||||||||
 |
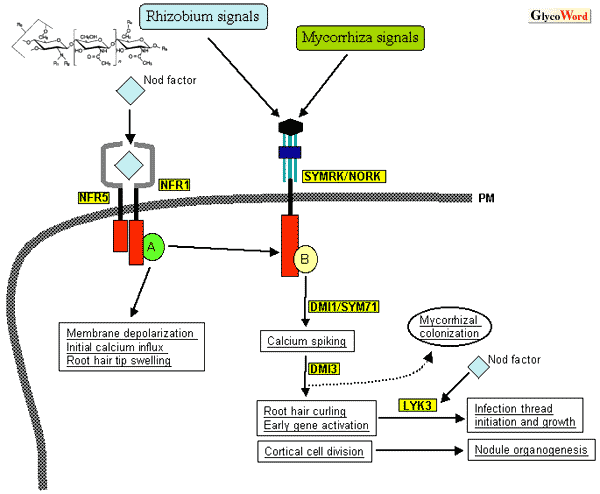
Nitrogen-fixing symbiosis of legume plants and rhizobium bacteria is initiated by recognition by the host plants of lipochitin oligosaccharides signal molecules (Nod factors) produced by rhizobium. We have previously described the structures of Nod factors, the chemical basis of the host specificity and their biological activities (“Interaction Between Leguminous Plants and Rhizobia Mediated by Nod Factors,” GlycoWord, 1999). Here we focus on recent progress made in the understanding of molecular mechanisms underlying Nod factor perception and subsequent signal transduction pathways. The earliest responses of legume plants to Nod factors appear on root hair cells, i.e., membrane depolarization, alkalization of extracellular fluid, and transient Ca++ influx into root hair cells, followed by oscillation of Ca++ influx/efflux (Ca-spiking). Concomitantly, root hairs show typical morphological changes, such as tip swelling, multiple tip growth (branching), and abundant deformation. These responses prepare the rhizobial infection process. Nod factors also elicit cortical cell division to form nodule primordium and activate nodule organogenesis. Two legume species, Lotus japonicus and Medicago truncatula, have been proposed as model legumes that are suitable for molecular genetic studies. Using these model legumes, host plant genes responsible for Nod factor perception and early signal transduction pathways were recently cloned in rapid succession. Ljsym1 and Ljsym5 are both nodulation-defective (nod-) mutants of L. japonicus and show no response to Nod factor application at all. Therefore, the responsive genes were postulated to be the Nod factor receptor itself or to be involved in the earliest steps of the Nod factor signaling cascade. Those genes were isolated by positional-cloning and termed NFR1 and NFR5, respectively. NFR1/5 both encode for membrane-spanning receptor protein kinase with the LysM domain in their putative extracellular structures. LysM is a domain involved in the substrate recognition of peptideglycan-degrading enzymes that cleave Another LysM receptor kinase LYK3 was cloned from M. truncatula. LYK3 was isolated as an ortholog of Pisum sativum SYM2 which is known to be required for progression of the infection process with a specific strain of rhizobium that produces Nod factors with specific modifications. Knock-down of LYK3 by RNAi technique demonstrated that the infection process stops after the infection thread initiation unless the rhizobial Nod factors have specific decorations. Rhizobium bacteria continue to secrete Nod factors even in the infection threads so the specific recognition of Nod factors by LysM receptor kinases also plays a role in the later stages of infection process. Besides legume-rhizobium symbiosis, symbiosis with arbuscular mycorrhizal fungi is distributed much more widely in the plant kingdom. Mycorrizal symbiosis was established evolutionally far before legume-rhizobium symbiosis. More than half of the nod- mutants of legumes identified so far are also defective in mycorrhizal symbiosis (myc-), indicating that both symbioses share partially overlapped genetic programs. The NFR1/5 mutants are both normal in mycrrhizal symbiosis (myc+), strongly suggesting that these mutations are involved in the earliest perception of Nod factors. On the other hand, most nod-myc- mutants exhibit some responses, if not the same as those in wild type plants, to Nod factors. SYMRK and NORK isolated from L. japonicus and M. truncatula, respectively, are receptor kinases with leucine-rich repeat (LRR) in their extracellular domain. Those mutants are nod-myc-, and show substantial root hair swelling and/or deformation upon Nod factor application. However, they show neither Ca-spiking nor activation of symbiosis-specific genes such as leghemoglobin. Thus SYMRK/NORK are postulated to play a role in the signaling cascade leading from the initial Nod factor perception with NFR1/5 to Ca-spiking and subsequent symbiosis-specific gene activation. In very recent years, a number of plant genes, such as SYM71/DMI1 and DMI3, which are presumably involved in early signaling pathways common to rhizobium and mycorrhizal symbioses have been cloned. Interestingly, the M. truncatula LYK3 gene is found to reside in a cluster of many similar genes including at least 7 LysM receptor kinase genes within a ca. 300kb DNA stretch. This is also the case in the L. japonicus NFR1 gene. It is well known that the strict host specificity of legume-rhizobium symbiosis is predominantly determined by the chemical structures of Nod factors, in particular various substitutions on the lipochitin backbone. To distinguish such structural diversity of Nod factors, the receptor gene must evolve rapidly within legume species. It is possible that the relatively high frequency of recombination within a cluster of similar gene copies ensured such rapid divergence of receptor genes, enabling very finely tuned recognition specificities during the legume evolution process. Molecular genetic studies with model legumes are now providing a long-awaited breakthrough for a better understanding of the molecular mechanisms of symbiotic plant-microbe interactions. Many plant genes that presumably play crucial roles in Nod factor perception and subsequent signaling pathways will be cloned in the coming few years. In the near future, it is expected that the functions and interrelationships of those gene products will solved by intense biochemical and molecular biological studies to uncover molecular mechanisms underlying plant-microbe symbiosis. |
|||||||||||||||||||
|
||||||||||||||||||||
| Hiroshi Kouchi and Yosuke Umehara (National Institute of Agrobiological Sciences) | ||||||||||||||||||||
|
||||||||||||||||||||
| Oct. 29, 2004 | ||||||||||||||||||||
|
|
||||||||||||||||||||
|
||||||||||||||||||||