

 |
 |
Fucosyltransferase Family |
||||||||||||||||||||||||||||||||||||||||||||||||||
 |
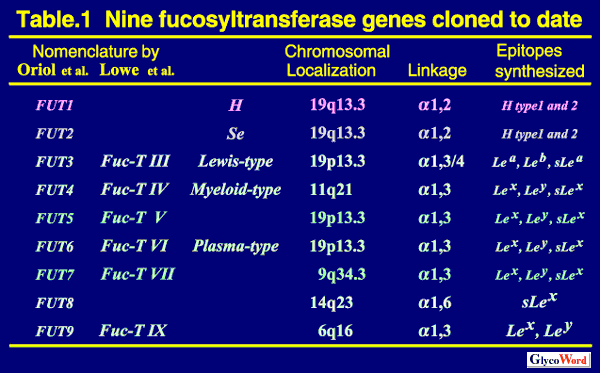
Two alpha1,2- and Five alpha1,3-fucosyltransferases Cloned to Date So far, nine human genes for two alpha 1,2-fucosyltransferases (alpha 1,2FUT), one arufa 1,6-fucosyltransferase (alpha1,6FUT) and six alpha 1,3-fucosyltransferases (alpha 1,3FUT) have been cloned. Their nomenclature, chromosomal localization and capacity for Lewis antigen synthesis are summarized in Table 1. |
|||||||||||||||||||||||||||||||||||||||||||||||||
 |
||||||||||||||||||||||||||||||||||||||||||||||||||
| FUT1 (H enzyme) and FUT2 (Se enzyme) are alpha 1,2FUTs catalyzing the transfer of fucose (Fuc) towards the galactose (Gal) residue of type 1 (Gal beta1,3GlcNAc) and type 2 (Gal beta1,4GlcNAc) chains, resulting in the synthesis of H-type 1 and H- type 2 chains, respectively. FUT1 (H) determines the expression of O-type antigen (H antigen) of the ABO blood group system on erythrocytes, whereas FUT2 (Se) determines it in saliva, i.e. secretor status. There is a very rare sporadic mutation of the H gene, and individuals with this mutation, referred to as Bombay or para-Bombay individuals, lack the H enzyme and the H antigens on erythrocytes (1). The levels of H antigen expression on erythrocytes of Bombay and para-Bombay individuals are determined solely by H enzyme activity. Individuals have been divided into two groups, secretors and nonsecretors, depending on the presence or absence of ABH antigens in their saliva. Nonsecretors were found to be homozygotes for the inactive FUT2 (Se) genes, which were inactivated by point mutations or crossover of the genes. The sej allele, which is an inactive Se allele and widely distributed among Asian people, occupies the frequency of 40% among Se alleles in the Japanese population. Thus, about 16% of the Japanese people are nonsecretors with the sej/sej genotype (1-3). The tissue distribution of H and Se enzymes has not yet been examined in detail. However, the epithelial cells of some digestive organs, i.e. colorectal and stomach tissues, are known to express both H and Se enzymes. | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||
|
FUT3 (Fuc-TIII) is the Lewis enzyme which determines the expression of Lewis blood group antigens on erythrocytes.The phenotypes of the Lewis blood group of erythrocytes are divided into four types, i.e. Le(a+b-), Le(a-b+), Le(a+b+) and Le(a-b-). The expression of Lewis blood group antigens on erythrocytes definitely requires the Lewis enzyme(Fig.1,Fig. 2) (4, 5). About 10% of the Japanese people genetically lack the Lewis enzyme and show the Le(a-b-) erythrocyte phenotype (5). They are homozygotes with the inactive Lewis (FUT3) alleles, i.e. with the Le/Le genotype. Three kinds of inactive Lewis alleles, which are inactivated by point mutations, were found in the Japanese population (3). The Le(a+b-) individuals are nonsecretors who completely lack the FUT2(Se) enzyme, but possess the active FUT3(Le) enzyme. The Le(a-b+) individuals possess both active Le and Se enzymes. The Le(a+b+) individuals possess the active Le enzyme, but their Se enzyme is weakened by point mutations. FUT3 is ubiquitously expressed in the epithelial cells of digestive organs, including mammary gland, lung, uterus etc. FUT3 exhibits both alpha 1,3 and alpha 1,4-fucosyltransferase activity, and is the only enzyme which is responsible for the type 1 Lewis antigens, such as Lea, Leb and sLea antigens, in a whole body (3, 6). The Le(a-b-) individuals, who are homozygotes of null Le alleles, cannot synthesize Lea, Leb and sLea antigens in the whole body. Cancer patients with the Le/Le genotype never show an elevated CA19-9 value, a well-known tumor marker detecting the amount of sLea antigens, but frequently show elevated DU-PAN-2 values, detecting the precursor structure lacking the fucosylation by FUT3 (3). FUT4, the myeloid-type alpha 1,3FUT, can synthesize Lex, Ley and sLex epitope, although the activity for sLex synthesis is very weak. FUT4 is ubiquitously expressed not only in myeloid tissue but also in various other tissues (7). The knock-out mouse lacking FUT4 looks healthy. FUT5 can synthesize Lex, Ley and sLex. However, it is still unclear which tissue expresses FUT5. FUT6 also can synthesize three type 1 Lewis antigens, Lex, Ley and sLex, and exhibits the strongest alpha1,3FUT activity among the five alpha 1,3FUTs. Many tissues redundantly express three alpha 1,3FUTs, i.e. FUT3, FUT4 and FUT6. FUT7 has strict specificity for substrates, and only synthesizes the sLex epitope. Tissue distribution of FUT7 is very restricted to leukocytes and high endothelial venule cells (8). In human native tissues, the sLex antigens, which is known to be the ligands for selectins, are synthesized mainly by FUT3 and FUT6 in cancer cells derived from gastrointestinal tissues (9), and by FUT7 in cells of myeloid lineage (10). FUT9 was most recently cloned among six alpha1,3FUTs (11-15). It is localized at human chromosome 6q16 (13). FUT9 exhibited very strong activity for the Lex and Ley synthesis, but not for the synthesis of sLex and any type 1 Lewis antigens. FUT9 is mainly expressed in neuronal and glial cells, proximal tubular cells in kidney, subglandular cells in stomach and granulocytes in peripheral blood cells (11, 12, 15). FUT9 preferentially fucosylates the distal GlcNAc residue of polylactosamine chain resulting in the synthesis of Lex epitope, while the other four alpha1,3FUTs (FUT3, FUT4, FUT5 and FUT6) preferentially fucosylate the inner GlcNAc residue resulting in the synthesis of internal Lex epitope (Fig. 3) (14, 15). FUT9 is a real Lex(CD15) synthase in vivo, while FUT4 is mainly involved in the CDw65 expression, not in the CD15 expression (14, 15). |
||||||||||||||||||||||||||||||||||||||||||||||||||
| Mark K. Narimatsu (Institute of Molecular and Cell Biology(IMCB), National Institutes of Advanced Industrial Science and Technology (AIST)) |
||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||
| Revised; May 15, 2001 | ||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||