|
Amino Acid Residues Essential for Transglycosylation Activity of Endo-ß-N-Acetylglucosaminidase
|
|
|
 |
Molecular structure of endo-ß-N-acetylglucosaminidase
Endo-ß-N-acetylglucosaminidase (endo-ß-GlcNAc-ase) releases N-linked oligosaccharide chains from glycoproteins by cleaving the di-N-acetylchitobiose (GlcNAc-GlcNAc) moiety unit. Endo-ß-GlcNAc-ase from Streptomyces plicatus (called Endo-H) has frequently been used as an important reagent in studies on the structure and physiological role of N-linked oligosaccharides. The Endo-H gene was cloned and sequenced in 1984, and the enzyme has been reported to share significant sequence homology with four endo-ß-GlcNAc-ases from Flavobacterium species (Endo-F1, Endo-F2, Endo- F3, and Endo-Fsp), and two regions of homology with chitinases from various origins. The three dimensional structure of Endo-H and Endo-F1 have been resolved by X-ray crystallography. These enzymes have highly conserved amino acid residues, especially the active site residues Asp130 and Glu132 (DXE) in Endo-H (Fig. 1). The glutamic acid has been identified as the proton donor, and the aspartic acid, stabilizing the intermediate in a substrate assisted hydrolysis mechanism in which the N-acetyl group of the substrate acts as the nucleophile (1). Moreover, comparison of the structure of Endo-F3 (specific for biantennary and triantennary complex oligosaccharides) with that of Endo-H (Endo-F1) reveals highly distinct folds and amino acid compositions at the oligosaccharide recognition sites (2). |
|
|
| Fig. 1 Schematic diagram of the proposed model for binding and recognition of the substrate by Endo-H |
|
|
|
|
|
Transglycosylation activity of endo-ß-GlcNAc-ase from Arthrobacter protophormiae (Endo-A)
Arthrobacter protophormiae, a gram-positive bacterium, produces endo-ß-GlcNAc-ase (called Endo-A) when the cells are grown in medium containing ovalbumin; the purification and properties of the enzyme have been reported. Endo-A hydrolyzed primarily high-mannose-type oligosaccharides but not complex-type oligosaccahrides. During a study on enzymatic kinetics, we found that exposing Endo-A to a mixture of monosaccharides such as glucose and mannose increased the apparent enzyme activity. Endo-A has transglycosylation activity, and the high-mannose-type oligosaccharides are transferred to suitable acceptors such as mono- and oligosaccharides (3). Endo-A could transfer high-mannose-type oligosaccharides not only to various glycosides but also to partially deglycosylated glycoproteins. Thus, Endo-A is useful for synthesizing neoglycoconjugates and for studying the biological functions of high-mannose-type oligosaccharides(4). |
|
|
|
|
|
Identification of amino acid residues essential for transglycosylation activity of Endo-A
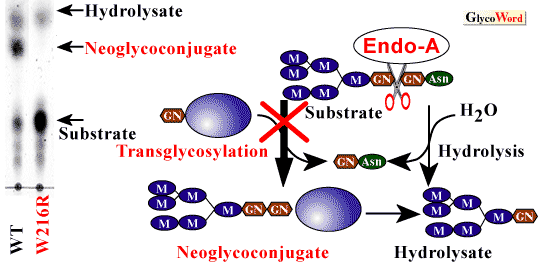
Using the degenerate oligonucleotides-encoding N-terminal and other determined sequences within Endo-A protein, the Endo-A gene was cloned. Endo-A consists of signal peptides of 24 amino acids and a mature protein of 621 amino acids (5). Endo-A has no sequence similarity with Endo-H, Endo-F1 or Endo-Fsp (these enzymes do not show transglycosylation activity). To determine which amino acids are involved in the transglycosylation acitivity of Endo-A, we employed PCR random and site-directed mutagenesis. We isolated several mutants, and these mutants had markedly reduced hydrolysis activity. Moreover, we isolated one mutant (W216R) that eliminated the transglycosylation activity without affecting the hydrolysis activity (6) (Fig. 2). It is difficult to explain why the transglycosylation activity is inhibited by the substitution of Trp216. However, the findings may prove important to the elucidation of the structure of Endo-A by X-ray crystallography, as well as to the definition of the catalytic mechanism of Endo-A and substrate recognition characteristics.
|
|
|
| Fig. 2 |
TLC analysis of Endo-A transglycosylation activity of mutant at position 216. Left, Man6GlcNAc2Asn was incubated with wild-type Endo-A or Mutant W216R in the presence of 0.1 M glucose for 10 min. Right, putative mechanism of Endo-A transglycosylation activity. |
|
 |
|
|
|
|
|
Endo-A homologues in various organisms
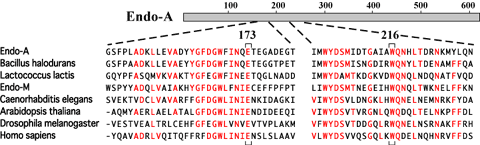
Recent database searches have revealed many homologues of Endo-A in various organisms. A gene for Endo-M, an endo-ß-GlcNAc-ase from Mucor hiemalis capable of transglycosylation of a complex biantennary oligosaccharide to peptidyl-GlcNAc (4), has been cloned and sequenced. Endo-M has 744 amino acids and 29% identity with the N-terminal region of Endo-A (6). We propose a new family of endo-ß-GlcNAc-ases, capable of catalyzing transglycosylation. In addition to Endo-M, the family includes putative endo-ß-GlcNAc-ases identified during the genome sequencing projects for Bacillus halodurans C-125 and Lactococcus lactis. Interestingly, similar proteins can also be also found in multiple eukaryotic organisms, such as worm (Caenorhabditis elegans), plant (Arabidopsis thaliana), fly (Drosophila melanogaster) and human (Homo sapiens)(Fig. 3). Thus, Endo-A seems to belong to a family that contains well-conserved members from prokaryotes to eukaryotic multicellular organisms. In particular, amino acids Glu-173 and Trp-216 in Endo-A, which were determined to be essential for catalytic activity, are completely conserved among these endo-ß-GlcNAc-ases. However, these entries in the database are all hypothetical proteins. It should be also noted that yeast species such as Saccharomyces cerevisiae and Schizosaccharomyces pombe do not possess any homologue among those whose genomes were completely sequenced. Future experiments will determine whether or not these Endo-A homologues have endoglycosidase (and transglycosylation) activity in the cell. Alternatively, Endo-A homologues might positively participate in various cellular functions in eukaryotic cells, such as degradation of misfolded glycoproteins in the ER. |
|
|
|
|
|
| Fig. 3 |
Amino acid sequence alignment of Endo-A homologues. Amino acids Glu-173 and Trp-216 in Endo-A, which were determined to be essential for catalytic activity, are indicated beneath the alignment.
|
|
 |
|
|
|
|
Kaoru Takegawa and Kiyotaka Fujita (Kagawa University, Faculty of Agriculture) |
|
|
|
| References |
(1) |
Rao V, Cui T, Guan C, Van Roey P : Mutations of endo-ß-N-acetylglucosaminidase H active site residues Asp130 and Glu132:Activities and conformations. Protein Sci. 8, 2338-2346, 1999 |
|
(2) |
Waddling CA, Plummer TH Jr., Tarentino AL, Van Roey P: Structural basis for the substrate specificity of endo-ß-N-acetylglucosaminidase F3. Biochemistry 39, 7878-7885, 2000 |
|
(3) |
Takegawa K, Yamaguchi S, Kondo A, Iwamoto H, Nakoshi M, Kato I, Iwahara S : Transglycosylation activity of endo-ß-N-acetylglucosaminidase from Arthrobacter protophormiae.Biochem. Int. 24, 849-855, 1991 |
|
(4) |
Yamamoto K, Takegawa K: Transglycosylation activity of microbial endoglycosidases and its application. Trends Glycosci. Glycotechnol. 48, 339-354, 1997. |
|
(5)
|
Takegawa K, Yamabe K, Fujita K, Tabuchi M, Mita M, Izu H,Waranabe A, Asada Y, Sano M, Kondo A, Kato I, Iwahara S: Cloning,sequencing, and expression of Arthrobacter protophormiae endo-ß-N-acetylglucosaminidase in Escherichi coli. Arch. Biochem.Biophys. 338, 22-28, 1997 |
|
(6)
|
Fujita K, Takegawa K : Tryptophan-216 is essential for the transglycosylation activity of endo-ß-N-acetylglucosaminidase A.Biochem. Biophys. Res. Commun. 283, 680-686, 2001 |
|
|
| Sep.15, 2001 |
|
|
|
|
|
|
|



