| Initiators for GAG biosynthsis |  |
|
 |
Proteoglycans (PG) are complex molecules which consist of a core protein with a number of covalently linked GAG chains. The linkage regions of chondroitin sulfate (CS), dermatan sulfate (DS), heparan sulfate (HS), and heparin (HP) chains, except keratan sulfate, to the polypeptide (PP) have a common structure, GlcA beta 1-3Gal beta 1-3Gal beta 1-4Xyl beta 1-O-PP. Biosynthesis of these GAGs is initiated by the transfer of Xyl from UDP-Xyl to a hydroxide group of specific Ser residues in PP. The transfer of Xyl to PP is essentially different from that of either GlcNAc or GalNAc to GlcA residue at the non-reducing terminal. The former is a reaction to determine the number of GAG chains in the proteoglycan ,1 and the latter determines the species of GAG.2 In other words, a specific signal which xylosyltransferase recognizes for the enzyme reaction is a determinant for the number of GAG chains. The species of GAGs is determined by either alpha GlcNAc T1 (transferase of GlcNAc to GlcA) or beta GalNAc T1 (transferase of GalNAc to GlcA) recognizing the formed PP with the specific signal. Furthermore, it has been suggested that aglycon (noncarbohydrate portion) of primer for initiation of GAG elongation regulates the length of GAG chains .3 Accordingly, this article describes the following contents, 1) recognition-signals for xylosyltransferase, 2) recognition-signals for alpha GlcNAc T1 and beta GalNAc T1, 3) aglycon-dependent means of controlling the length of GAG chains.
1) How is the specific Ser residues to be transferred Xyl determined ?
A study of comparisons of amino acid sequences of PG-core proteins whose cDNAs had been cloned assumed that some minimum peptide sequence was required as a substrate of the xylosyltransferase. Several mutants of the peptides that were changed some amino acid to another were generated, and Km and Vmax values of each peptide were measured. The optimal amino acid sequence as a substrate of the enzyme was determined according to Vmax/Km values and supposed as a recognition domain of the core protein for the enzyme. Based on the results, cDNA mutants of the core proteins were prepared and examined on each expression. Glu/Asp-X-Ser-Gly (X is any amino acid) was proposed to be a common amino acid sequence to be recognized by the enzyme.
2) How is the molecular species of GAGs determined ?
An experiment to answer the question was performed using betaglycan, syndecan, and glypican, which are hybrid type proteoglycans carrying both CS and HS chains on a single core protein, as model molecules. Several site-directed mutations were performed on a consensus amino acid sequence adjacent to GAG attachment sites found in the three proteoglycans. The cDNA mutants were transfected into CHO cells to examine the generated species and yield of GAG.2 The results indicated that an elongation of HS chains requires a cluster of acidic amino acids consisting of Glu and Asp in the distance of several amino acids from C-terminal of Ser-Gly dipeptide. Furthermore, it was shown that a hydrophobic amino acid such as Trp is required adjacent to the Ser-Gly. Interestingly, it was found that beta -D-Xylosides, artificial initiators for GAG synthesis, require a hydrophobic aglycon to initiate HS synthesis, being consistent with the requirement of hydrophobic amino acid.4 In a summary, it was suggested that the substrate-recognition site of alfa GlcNAc T1 enzyme to initiate HS synthesis consists of a linkage region of GlcA-Gal-Gal-Xyl, a hydrophobic amino acid adjacent to the Ser-Gly, and a cluster of acidic amino acids on the C-terminal part of Ser-Gly. On the other hand, the substrate-specificity of beta GalNAc T1 is still not clear regarding initiation of CS synthesis.
3) Effect of aglycon of artificial initiators of GAG synthesis, beta-D-Xylosides, on the length of GAG chains.
The mechanism to controlling the length of GAG chains is still not clear. However, it was reported that the length of GAG chains is related to the velocity of GAG synthesis, using the artificial initiators, beta-D-Xylosides.3 In Fig. 1, beta-D-Xylosides with various hydrophobicities were prepared, and incubated with cartilaginous tissues to examine the synthetic velocity of GAG elongated from the artificial initiators (Fig. 1A) and the length of GAG chains (Fig. 1B). The results indicated that the increase of hydrophobicities of the aglycons is related to the increase in the synthetic velocity of CS chains, resulting in the decrease in the length of synthesized CS chains. Thus, it was strongly suggested that an interaction between the aglycon portion and membrane of Golgi aparatus where GAG is synthesized influences the synthetic velocity of GAG, and that the length of GAG is controlled by the synthetic velocity.
| |
|
| Fig. 1 Effect of various beta-D-Xyls on synthetic velocity and chain length of chondroitin sulfate. |
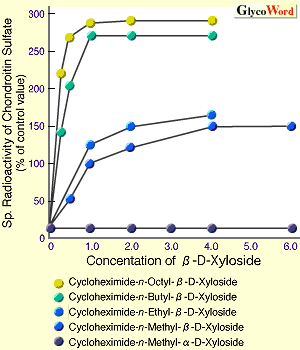 | | 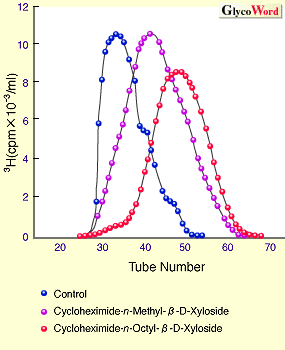 |
| A. Effect of various aglycons of beta-D-Xyloside on synthetic velocity of chondroitin sulfate in chick embryo-cartilaginous tissue. | | B. Comparison of patterns of gel filtration chromatrography (Sephadex G-200) of chondroitin sulfate chains generated from beta-D-xyloside with various aglycons. |
|
|
| Minoru Okayama (Kyoto Sangyo University, Faculty of Engineering) | |
|
|
| References | (1) | Bourden , MA, Extracellular Matrix Genes(eds. L.J.Sandell, C.D.Boyd), pp157-174, Academic Press, New York,1990 |
| (2) | Zhang, L, Esko, JD, J. Biol. Chem. 269, 19295-19299, 1994 |
| (3) | Robinson, HC, Brett, MJ, Tralaggan, PJ, Lowther,DA,and Okayama,M, Biochem. J. 148, 25-34, 1975 |
| (4) | Fritz, TA, Lugemwa, FN, Sarkar, AK, Esko, JD, J. Biol. Chem. 269, 300-307, 1994 |
| | |
| |
|
| Jun.15, 1998 |
|
| |
|
|
|
|



