

 |
 |
Glycosaminoglycan Sulfotransferase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 |
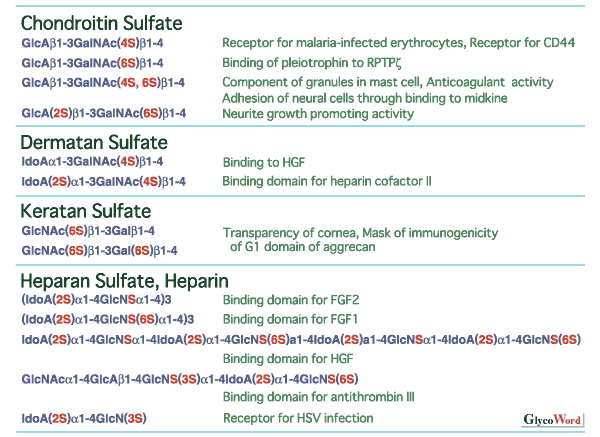
Divergent structure and sulfation of glycosaminoglycans Sulfated glycosaminoglycans are classified into chondroitin sulfate, dermatan sulfate, keratan sulfate, heparan sulfate and heparin by the structure of the repeating disaccharides. Sulfated glycosaminoglycans show marked structural diversity due to the sulfation pattern of the repeating disaccharides as well the backbone structures. As shown in Fig.1, the structural diversity of glycosaminoglycans correlates with various biological functions. Various sulfotransferases with strict substrate specificities are involved in the sulfation, a key step for the formation of the divergent structures. Each glycosaminoglycan sulfotransferase transfers sulfate to the defined position of the sugar residues from active sulfate (3'-phosphoadenosine 5'-phosphosulfate, PAPS). Some sulfotransferases are known to recognize not only the kind and position of acceptor sugar residues but also the structure of the neighboring sugar residues. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Substrate specificities of sulfotransferases involved in the biosynthesis of glycosaminoglycans In Table 1, possible substrate specificities of various sulfotransferases, which transfer sulfate to repeating disaccharide moieties, are listed with serial numbers. Most of these sulfotransferases have been so far purified and/or cloned. From the studies on the substrate specificities of the purified and/or cloned sulfotransferases, each sulfotransferase transfers sulfate to the defined position of the defined sugar residue. However, some sulfotransferase showed some ambiguity in acceptor substrate specificity (acceptor specificity was indicated in the following description by the serial number listed in Table 1): C6ST showed number 10 activity besides number 1 activity; and HS2ST showed not only number 17 activity but also number 18 activity. Several sulfotransferases that transfer sulfate to position 6 of nonreducing terminal GlcNAc residue have been cloned. Of these sulfotransferases, C-GlcNAc 6ST gene was identified as a gene that was inactivated in macular corneal dystrophy. Sulfation of keratan sulfate was decreased in the cornea of patients of macular corneal dystrophy; therefore, C-GlcNAc 6ST is a most likely candidate for GlcNAc 6ST involved in the synthesis of keratan sulfate in the cornea. Every NDST isoforms were shown to be bifunctional enzymes containing both number 13 activity and N-deacetylase activity. The carboxyl-terminal half of NDST-2 was able to catalyze number 13 activity (38). Sulfotransferases often recognize not only the sugar residues to which sulfate is transferred but also the structure of the neighboring sugar residues. For examples, HS2ST transfers sulfate to position 2 of IdoA adjacent to nonreducing side of GlcNS, but does not transfer sulfate to IdoA adjacent to nonreducing side of GlcNS(6SO4); 3-O-ST-1 and 3-O-ST-2 transfer sulfate to position 3 of GlcNS residues adjacent to reducing side of GlcA and IdoA(2SO4) or GlcA(2SO4), respectively, and 3-O-ST-3A transfers sulfate to position 3 of GlcN residues adjacent to IdoA(2SO4); and HS6ST-1 and HS6ST -2 preferentially transfer sulfate to position 6 of GlcNS residues adjacent to reducing side of IdoA and GlcA, respectively. Sulfotransferase genes Glycosaminoglycan sulfotransferases so far cloned has revealed to be type II transmembrane proteins except for 3-O-ST-1, which was assumed to be an intralumenal protein. From the homology observed in the deduced amino acid sequence, these sulfotransferases are classified to five groups indicated in Table 2. No significant homology was observed between each group. Sd protein and Pipe ST2 were gene products that are involved in chromosome segregation and in the determination of dorsal-ventral polarity, respectively, in Drosophila. Although these gene products show a significant homology to HS2ST, it has not been confirmed yet whether these gene products possess sulfotransferase activity. Topedo NSIST was cloned by expression-cloning using monoclonal antibody that recognizes a carbohydrate-epitope expressed on dystroglycan and other constituents of Torpedo electric organ synaptic membranes. NSIST shows relatively high homology to C6ST and the expressed protein showed C6ST activity in vitro (unpublished observation). In C6ST group and C4ST group, in addition to glycosaminoglycan sulfotransferases, glycoprotein sulfotransferases, which are involved in the sulfation of sugar chains attached to glycoproteins, are also included. Sulfation of sugar chains of glycoprotein and sulfation of glycosaminoglycans are assumed to be catalyzed by the enzymes evolved from a common ancestral protein. PAPS binding sites in sulfotransferases Two putative PAPS binding sites were found by X-ray crystallography of estrogen sulfotransferase; one is PKSGTTW for 5’-phosphate binding site (5'PSB) and another is X4RX7SX3 for 3'-phosphate binding site (3'PB) (Fig. 2) (43). The second amino acid, K (R), and the fifth amino acid, T (S), in the 5’PSB, and R and S in 3’PB indicated by bold letters in Fig. 3 were well conserved in almost sulfotransferase so far cloned except for NST (Fig. 3). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Osami Habuchi (Aichi University of Education) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| May 15, 2001 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||