| Biosynthesis and Degradation of Heparan Sulfate | |
|
 |
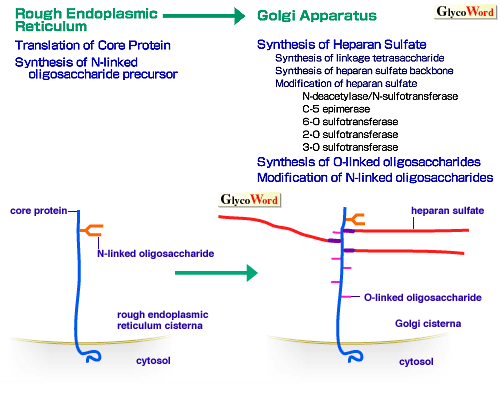
Heparan sulfate is synthesized as the glycosaminoglycan component of heparan sulfate proteoglycans. It is widely distributed on cell surfaces and basement membranes in mammals. It displays specific interactions with many biologically active proteins and, thus, is involved in many important biological processes (see A00-A02 of this series). Fig. 1 shows diagrams depicting biosynthesis of heparan sulfate proteoglycans and intracellular degradation steps of cell surface heparan sulfate proteoglycans (1). Heparan sulfate is synthesized in the Golgi apparatus, on the linkage tetrasaccharide [GlcA-Gal-Gal-Xyl-(Ser)] structure built on the serine residues in the core protein (A06 of this series), and contains repeating (typically 40-100 times) disaccharides [GlcA-GlcNAc]n. The backbone of the heparan sulfate chain appears to be synthesized by an enzyme ('heparan sulfate synthase'), which possesses both glucuronosyltransferase and N-acetylglucosaminyltransferase activities (2). This basic sugar structure is modified by a series of sulfotransferases and carbohydrate-modifying enzymes, including N-deacetylase/N-sulfotransferase (an enzyme which has two activities), C-5 epimerase, 2-O sulfotransferase, 6-O sulfotransferase, 3-O sulfotransferase. These modifications occur non-uniformly throughout the heparan sulfate chain, thus forming complex sulfation patterns with the concomitant presence of highly modified domains and unmodified domains (2). Specific sulfation patterns are critically important for the binding of heparan sulfate chains to proteins (see A01of this series). | |
|
| Fig. 1A Biosynthetic processes of heparan sulfate proteoglycans in rough endoplasmic reticulum and Golgi apparatus. |
|
 |
|
| Fig. 1B Intracellular degradation of cell surface heparan sulfate proteoglycans. |
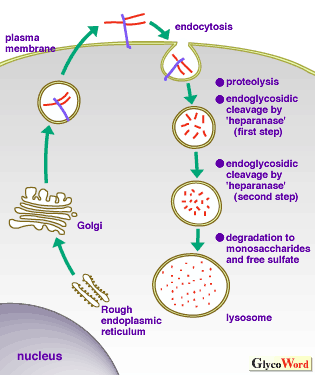
After endocytosis, heparan sulfate proteoglycans undergo partial degradation by closely coordinated proteolytic and endoglycosidic degradations in an environment with a neutral pH. In the next stage, in an acid environment, heparan sulfate fragments undergo further endoglycosidic degradation. The final stage of degradation involving exoglycosidases and sulfatase occurs in the lysosome and totally digests heparan sulfate into monosaccharides and free sulfate. See ref. 1 for details. |
|
|
|
How can the biosynthesis of heparan sulfate, which involves multiple carbohydrate-synthesizing enzymes, be regulated? This is where the regulatory mechanisms for biosynthesis of complex carbohydrates, including heparan sulfate, differ significantly from those of protein molecules. The latter can be regulated primarily by controlling gene transcription and translation mechanisms. On the other hand, regulation of complex carbohydrate biosynthesis must include many cellular functions including (a) regulation of acceptor protein (core protein) biosynthesis, (b) regulation of multiple glycosyltransferases, sulfotransferases, epimerase, etc., each requiring separate biosynthetic regulation, (c) integration of complex carbohydrate biosynthetic machinery through modulation of organella functions in rough endoplasmic reticulum and in Golgi apparatus. In order to account for the unique structure/function relationship of heparan sulfate, an even more complex regulatory scheme emerges. It boils down to the question how can specific sulfation patterns of heparan sulfate be synthesized (2,3)? Regulation of sulfotransferase activity alone would only increase overall sulfation of the molecule, but might not attain an efficient formation of desired, biologically active sulfation patterns. Functional templates (e. g., enzyme complexes to generate specific patterns of sulfation) can be imagined. Also, the localization of carbohydrate- synthesizing enzymes in the Golgi apparatus is an important factor that would certainly affect the biosynthetic regulation of heparan sulfate proteoglycans (4). How the genetic regulation of multiple carbohydrate-synthesizing enzymes can be conducted is also an interesting question. The regulation of the biological functions of heparan sulfate proteoglycans does not end at biosynthetic processes. Transportation of the molecules to their destination, i.e., the plasma membrane, basement membrane, etc., and their catabolic pathways are equally important. The cellular metabolism of cell surface heparan sulfate proteoglycans shows unique features as summarized in Fig. 1. The majority of cell surface heparan sulfate proteoglycans are endocytosed and undergo unique prelysosomal and classic lysosomal degradations. Thus, most mammalian cells contain substantial amounts of heparan sulfate degradation intermediates. It would be interesting to explore what their functions are. The processes involved in the metabolic turnover of heparan sulfate proteoglycans in extracellular matrix are currently largely unknown. Considering the biological roles of heparan sulfate proteoglycans in this location, i.e., integration of matrix structure, modulation of growth factor activities, etc., elucidation of their metabolic regulation poses a very interesting question.
| |
|
| Masaki Yanagishita (Tokyo Medical and Dental University ) | |
|
|
| References | (1) | Yanagishita, M : Cellular catabolism of heparan sulfate proteoglycans. TIGG 10, 57-63 ,1998 |
| (2) | Lindahl, U, Kusche-Gullberg, M, Kjellen, L : Regulated Diversity of Heparan Sulfate. J. Biol. Chem. 273, 24979-24982 ,1998 |
| (3) | Rosenberg, RD, Shworak, NW, Liu, J, Schwartz, JJ, Zhang, L : Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? J. Clin. Invest. 100 (11 Suppl.) : S67-75, 1997 |
| (4) | Uhlin-Hansen, L, Yanagishita, M : Differential effect of brefeldin A on the biosynthesis of heparan sulfate and chondroitin/dermatan sulfate proteoglycans in rat ovarian granulosa cells in culture. J. Biol. Chem. 268, 17370-17376, 1993 |
| | |
| |
|
| Dec. 15, 1998 |
|
| |
|
|
|
|




