|
Glycoside Hydrolase (GH) is a large class of enzymes classified under 3.2.1.- in the EC numbering system. In the Carbohydrate-Active enZymes (CAZy) database, GHs are classified into more than 160 families and consist of numerous enzymes including amylases and cellulases. The catalytic center (catalytic residues) of GH generally consists of a pair of aspartic acid (Asp) or glutamic acid (Glu). In exceptional cases, several GH families have been known to utilize a reaction mechanism involving a group in the substrate (N-acetyl or hydroxy group), and very few enzymes utilize a tyrosine or histidine as the catalytic residue. However, the catalytic residues of GH are basically the same in a variety of families.
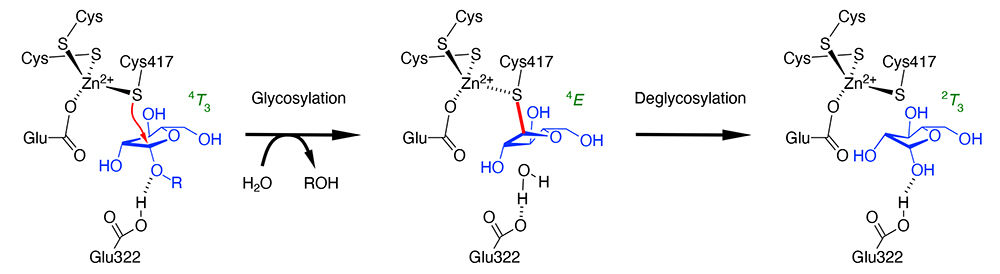
The authors found in 2014 that β-ʟ-arabinofuranosidase (HypBA1) from Bifidobacterium longum has the catalytic center with three cysteines and one glutamic acid coordinating to a zinc ion, one of which (Cys417) was near the anomeric carbon (C1 atom) of arabinofuranose in the enzyme (1). Since HypBA1 has been shown to catalyze anomer-retaining hydrolysis, this fact strongly suggested that the cysteine acts as the catalytic nucleophile. In other words, Cys417 was expected to act like Asp and Glu as the nucleophile residue in normal anomer-retaining GH. Glu322 is hydrogen bonded to the O1 hydroxy group of arabinofuranose, which is expected to function as the catalytic acid/base residue as in normal anomer-retaining GH. However, there have long been numerous studies that cysteine was suggested as the catalytic residue of GH, based on experimental results that cysteine modifiers cause loss of enzyme activity, all of which were later denied and found to have Asp or Glu as the catalytic residues. Therefore, when an unprecedented amino acid residue was predicted to be the catalytic center, as in the case of HypBA1, further verification was needed to prove the proposed reaction mechanism. Another question raised was why the deglycosylation reaction occurs once the covalent intermediate has been formed, because of the high stability of cysteine glycoside.
Two main approaches were adopted to validate the reaction mechanism of HypBA1 (2,3). These were the use of inhibitors as probes specifically labeling the cysteine residue, and the introduction of computational chemistry methods. As specific inhibitors, we used haloacetamide compounds with an arabinofuranosyl group and a cyclophellitol compound in the configuration of β-ʟ-arabinofuranose. The former has been used successfully in labeling peptide: N-glycanase, whose catalytic nucleophile residue is a cysteine, and the latter has been used as a specific inhibitor for a number of GH enzymes. X-ray crystallography of HypBA1 reacted with these inhibitors revealed that both compounds form covalently bound adducts to Cys417 in the crystal structures. These are mimics of the reaction intermediate. Together with the previously obtained arabinofuranose complex structure (corresponding to the reaction product) and the complex structure with the synthetic substrate p-nitrophenyl-β-ʟ-arabinofuranoside (corresponding to the Michaelis complex), three crystal structures along with the total hydrolysis pathway were obtained: before reaction, intermediate, and after reaction (Figure). The crystal structures provided information on the conformation of the five-membered ring of arabinofuranose, which changes along the reaction pathway (4T3 → 4E → 2T3). As for the problem in deglycosylation, detailed QM/MM calculations revealed that the reaction proceeds through a transition state that is reasonable in terms of free energy changes. It was suggested that the energetics of the deglycosylation reaction is finely tuned by the Zn2+-Cys417 interaction. HypBA1 belongs to GH127, and enzymes of another family (GH146) are also known to have the catalytic center consisting of three cysteines and one glutamic acid coordinating to a zinc ion. More GH enzymes with cysteine as the catalytic residue will be possibly found in the future.
Shinya Fushinobu
(Department of Biotechnology, The University of Tokyo)
| References |
| (1) |
Ito T, Saikawa K, Kim S, Fujita K, Ishiwata A, Kaeothip S, Arakawa T, Wakagi T, Beckham GT, Ito Y, Fushinobu S: Crystal structure of glycoside hydrolase family 127 β-L-arabinofuranosidase from Bifidobacterium longum. Biochem. Biophys. Res. Commun. 447, 32-37, 2014 |
| (2) |
McGregor NGS, Coines J, Borlandelli V, Amaki S, Artola M, Nin-Hill A, Linzel D, Yamada C, Arakawa T, Ishiwata A, Ito Y, van der Marel GA, Codée JDC, Fushinobu S, Overkleeft HS, Rovira C, Davies GJ: Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent β-L-Arabinofuranosidase Inhibitor. Angew. Chem. Int. Ed. 60, 5754-5758, 2021 |
| (3) |
Maruyama S, Sawano K, Amaki S, Suzuki T, Narita S, Kimura K, Arakawa T, Yamada C, Ito Y, Dohmae N, Fujita K, Ishiwata A, Fushinobu S: Substrate complex structure, active site labeling and catalytic role of the zinc ion in cysteine glycosidase. Glycobiology 32, 171-180, 2022 |
Jun. 15, 2023
|
|---|