
Paul H. Weigel
Paul Weigel, Ph.D., obtained a B.A. degree in Chemistry from Cornell University and his doctoral degree from The Johns Hopkins University School of Medicine in 1975. He joined the University of Texas Medical Branch at Galveston as an Assistant Professor in 1978 and became a Professor of Biochemistry and Cell Biology in 1987. Since 1994 he has been Professor and Chairman in the Department of Biochemistry & Molecular Biology, College of Medicine, at the University of Oklahoma Health Sciences Center in Oklahoma City. Dr. Weigel has made substantial research contributions in several fields, including the discoveries of multiple coated pit pathways for receptor-mediated endocytosis and of the enzyme responsible for hyaluronan biosynthesis. He has a long-standing interest in the biochemistry and the biology of hyaluronan. His group was the first to identify and isolate the gene for hyaluronan synthase. Dr. Weigel’s laboratory also developed the use of structurally defined iodinated hyaluronan oligosaccharides of high specific radioactivity to detect and to study specific hyaluronan receptors and binding proteins. This approach enabled his laboratory to purify and then clone the rat and human liver endothelial cell receptors that remove circulating hyaluronan and other glycosaminoglycans from the blood by receptor mediated endocytosis. This membrane receptor has several designations, including HARE, the Hyaluronan Receptor for Endocytosis.
The initial article (in 1998) on bacterial hyaluronan synthases in this series1 discussed what was known about the enzymes responsible for hyaluronan synthesis. After our discovery in 1993 of the first gene encoding an hyaluronan synthase from Group A Streptococcus, we and many others identified similar hyaluronan synthase genes (or cDNAs) in other bacteria and a wide range of eukaryotes2-6. All but one of these >20 hyaluronan synthases comprise a family of proteins with common structural and mechanistic features. During the past five years, substantial progress has been made toward understanding the structure and function of the bacterial hyaluronan synthases and how they produce hyaluronan.
We now know of four types of bacteria that produce hyaluronan capsules as part of their pathogenic arsenal for infecting various animal hosts. Bacteria that coat themselves with a thick layer of hyaluronan are able to “ hide” from the immune systems of their host. These bacteria are Streptococcus equisimilis (a pathogen in animals and sometimes humans), Streptococcus pyogenes (a human pathogen), Streptococcus uberis (a bovine pathogen), and Pasteurella multocida (a fowl pathogen). Based on their similarities and differences, the known hyaluronan synthase proteins have been divided into two categories, designated Class I and Class II6. The streptococcal hyaluronan synthases are Class I family members, whereas the hyaluronan synthase from Pasteurella multocida (pmHAS) is the only Class II member7.
The Class II pmHAS enzyme is very different than the Class I hyaluronan synthases in its structure, membrane topology, and mechanism of hyaluronan synthesis (Table 1). The major structural difference between Class I and Class II synthases is the overall organization and distribution of membrane domains within the two types of proteins (Fig. 1). The Class I family members have the same topological organization within their first ~420 amino acids, although the topology has been experimentally determined only for the spHAS8. The Class I enzymes contain 6-8 transmembrane or membrane-associated domains distributed throughout the protein - from near the amino terminus to near the carboxyl terminus.
Table 1 Structural and mechanistic differences between the Class I and Class II hyaluronan synthases.
| Class I | Class II | |
|
|
||
| Family Members | Over 20 members cloned (e.g. Streptococcus, avian, amphibian, mammalian) | One member (P. multocida) |
|
|
||
| Size (amino acids) | 417-588 | 972 |
|
|
||
| Membrane attachment of the protein | Six to eight membrane-associated domains from N- to C-terminus | C-terminal membrane anchor |
|
|
||
| Active as a truncated soluble protein | No | Yes (e.g. residues 1-703) |
|
|
||
| Hyaluronan chain growth | At reducing end | At nonreducing end |
|
|
||
| Ability to elongate primer oligosaccharides | No evidence for extension of hyaluronan oligosaccharides | Readily extends hyaluronan oligosaccharides |
In contrast, the Class II enzyme has a membrane attachment domain very near the carboxyl terminus that anchors the protein by an unknown mechanism; probably by interaction with another membrane-anchored component. The major mechanistic difference is that the two Classes of synthases extend hyaluronan at opposite ends of the polysaccharide. The Class II pmHAS adds new sugars to the nonreducing end, whereas the Class I enzymes extend at the reducing end. Unlike the Class I enzymes, the Class II pmHAS has a two-domain modular structure with two transferase activities that alternatively bind and release hyaluronan chains to add new sugars to the nonreducing end by “classical” glycosyltransferase mechanisms. The rest of the Update will focus on new information about the Class I synthases.
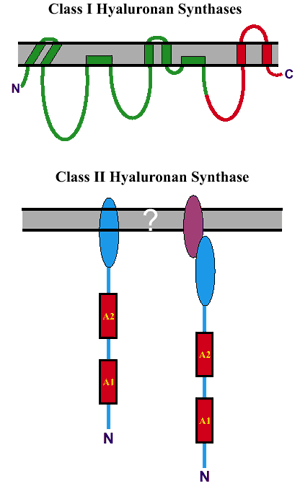
Fig. 1 Topological differences between Class I and Class II hyaluronan synthases.
Top: The streptococcal Class I synthases (green line) contain four transmembrane domains and two membrane-associated domains8. The eukaryotic Class I synthases are larger and predicted to contain two additional transmembrane domains (red line). Bottom: The Class II hyaluronan synthase is membrane-associated via an unknown mechanism; either directly due to a membrane-associated domain at the C-terminus of the native pmHAS (bottom left), or indirectly due to a binding interaction between a C-terminal domain and an unidentified integral membrane component (bottom right).
A. Overview.
Hyaluronan synthases are membrane-bound enzymes in bacteria and eukaryotic cells, and active hyaluronan synthases are found in the cell or plasma membrane. Recent findings of intracellular hyaluronan9 indicate that active hyaluronan synthases may also be present inside cells. The hyaluronan synthases are unusual in that these proteins have two different glycosyltransferase activities within the same protein. Furthermore, after each sugar addition, the new product hyaluronan chain becomes the substrate for the next sugar addition. Historically, the net reaction describing the assembly of one disaccharide unit onto a hyaluronan chain has been presented as the addition of new sugars at the nonreducing end. In fact, this is only one of the two possible reaction mechanisms. The second reaction mechanism (Fig. 2A) describes addition of a disaccharide unit at the reducing end of the polymer. In this mechanism, the hyaluronan chain is attached to UDP at its growing end.
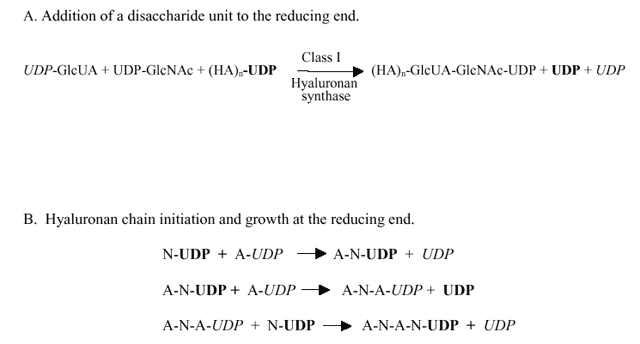
Fig. 2 The mechanism of hyaluronan synthesis at the reducing end.
GlcUA and GlcNAc, are indicated by A and N, respectively, and the UDP groups for their UDP-sugars are indicated, respectively, in italic or boldface font. The number of disaccharide units is indicated by n. The UDP-sugar added to the polymer chain is transferred intact, without cleavage of its UDP linkage. The UDP released during each transfer step comes from the HA-UDP intermediate formed by the addition of the previous sugar. Thus, of the two net UDP groups released when a disaccharide unit is assembled at the reducing end (A), only one UDP comes from the last two UDP-sugars added. The other UDP comes from the hyaluronan-UDP chain; from the last sugar added prior to addition of the new disaccharide unit. A; the addition of two sugars (a disaccharide unit) at the reducing end of a hyaluronan chain. B; the initiation and early extension of a hyaluronan chain at the reducing end.
B. Multiple functions of Class I hyaluronan synthases.
The two activated sugar precursors for hyaluronan biosynthesis are UDP-glucuronic acid (UDP-GlcUA) and UDP-N-acetylglucosamine (UDP-GlcNAc). In order to make a disaccharide unit and extend the growing hyaluronan chain, Class I hyaluronan synthases possess at least six distinct functions (Fig. 3). These activities include two different sugar nucleotide binding sites, two different glycosyltransferase activities, one or more binding sites for the growing hyaluronan-UDP chain and the ability to transfer the hyaluronan chain, within the enzyme, to set up the next round of saccharide addition. The UDP-sugar substrates are produced and used by the hyaluronan synthase inside the cell8, and the hyaluronan chain is continuously transferred (translocated) across the membrane so that it is extruded to the cell exterior. In the case of bacteria, this hyaluronan forms a capsule, whereas for many eukaryotic cells, the result is either that hyaluronan forms a pericellular coat surrounding the cell or is released into the surrounding extracellular matrix.
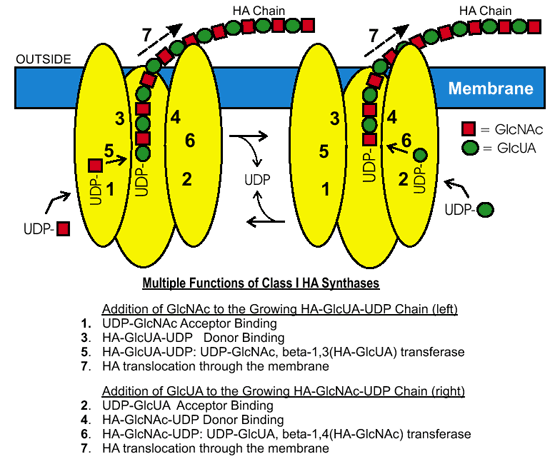
Fig. 3 Class I enzymes need multiple functions for hyaluronan biosynthesis.
The diagram shows a membrane-bound Class I hyaluronan synthase and the individual activities required for the enzyme to make a disaccharide unit and extend the growing hyaluronan chain at the reducing end. The nucleodtide-sugar substrates are produced and used by the synthase inside the cell, and the hyaluronan chain is continuously transferred (translocated) to the exterior of the cell. Two enzymes are shown to illustrate how one enzyme could alternately make each of the two types of glycoside bonds in hyaluronan; a similar alternate model is possible in which all substrates are bound and a disaccharide unit is added in a coordinated manner.
C. The Class I hyaluronan synthases polymerize hyaluronan by addition to the reducing end.
(i) Background. Two previous studies using membrane preparations from eukaryotic cells10-11 suggested that hyaluronan synthesis occurs at the reducing end. Since membranes contain other glycosyltransferase activities, however, these results might have alternate interpretations. Also, possible separate contributions of the three mammalian hyaluronan synthase isoenzymes to these results were unknown. In order to confirm this important conclusion, we performed similar experiments using two different purified, recombinant streptococcal hyaluronan synthases. Our results verify that addition of new saccharide units does, in fact, occur at the reducing end. We performed two types of experiments to determine direction of synthesis. Here I will summarize some of the evidence from these studies, which have been described in preliminary form12 and will be presented in detail elsewhere.
(ii) Pulse-labeling Approach. In the first series of experiments, we used a well-established strategy11,13 in which growing hyaluronan chains were pulse-labeled either early or late during a single round of chain synthesis. These radiolabeled hyaluronan products were then treated with two exoglycosidases (i.e. β-N-acetylglucosaminidase and β-glucuronidase) that act only on the nonreducing end to release one sugar at a time from individual hyaluronan chains. The results using either purified spHAS or seHAS showed that the first sugars incorporated into hyaluronan chains were more readily released than the last sugars incorporated, i.e. the earliest sugars incorporated into hyaluronan are closest to the nonreducing end.
For example, if hyaluronan produced by seHAS was briefly labeled with UDP-[3H]GlcNAc and then incubated with nonlabeled substrate, 48% of the radiolabel was subsequently released after 120 min of glycosidase treatment (Fig. 4). In contrast, only 11% of the radioactivity was released when the radiolabeling order was switched, so that the last sugars to be added to the growing hyaluronan chains were radiolabeled. The same results were obtained using both UDP-sugars and both streptococcal HASs. Since the latest sugars added must be closest to the growing end of the polymer, this is the reducing end.
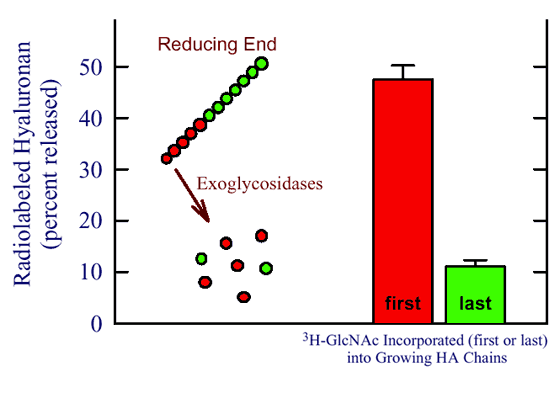
Fig. 4 Class I synthases add sugars at the reducing end of hyaluronan.
Hyaluronan chains were pulse labeled with UDP-[3H]GlcNAc either at the beginning (first; red) or the end (last; green) of one round of chain synthesis by the purified seHAS. The radiolabeled hyaluronan was then treated for 2 hours with exoglycosidases that remove sugars only from the nonreducing end and released radioactivity was determined. The cartoon (left) illustrates the distribution of labeled GlcNAc in a hyaluronan chain in each of the two labeling situations. The prediction is that the first sugars added will be the most readily removed by glycosidases only if they are added at the reducing end. As the growing chain gets larger the last sugars are farther away from the nonreducing end and less likely to be released by the glycosidases.
(iii) Demonstration of a dynamic hyaluronan-UDP lingkage. Our second strategy to determine direction of synthesis was to show that UDP is covalently attached at the reducing end of growing hyaluronan chains and that this linkage is dynamically broken and reformed as synthesis proceeds. Using purified seHAS and β-32P-labeled UDP-GlcNAc, we found consistent and reproducible incorporation of 32P into hyaluronan (Fig. 5). Since only the terminal sugar unit in a hyaluronyl-GlcNAc-[32P]-P-Uridine molecule is labeled, the amount of 32P-HA recovered in such experiments is relatively low, e.g. compared to labeling 50% of all the sugars in hyaluronan chains when using UDP-[3H]GlcNAc.
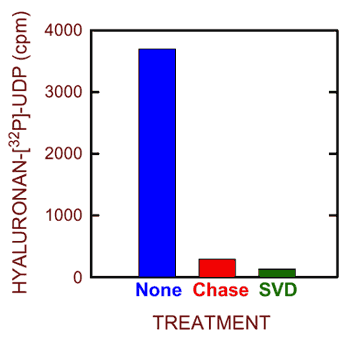
Fig. 5 Synthesis of hyaluronan-UDP by purified seHAS.
Purified seHAS was incubated for 1.5 min at 25°C in the presence of unlabeled UDP-GlcUA and [32P]UDP-GlcNAc. Parallel samples were then treated with nothing (blue), “chased” with an excess of each unlabeled UDP-sugar (red) or treated with snake venom phosphodiesterase (green) and the incorporation of 32P into hyaluronan was assessed by paper chromatography. The background in the absence of enzyme has been subtracted.
Nonetheless, radiolabeling of hyaluronan with 32P was readily detected. The conclusion that this 32P-hyaluronan reflects the presence of UDP covalently attached at the reducing end is supported by a variety of controls. For example, the labeled material was destroyed by treatment with hyaluronidase or snake venom phosphodiesterase (Fig. 5). Most importantly, the radiolabel was removed by the addition of unlabeled UDP-sugars. This “chase” control confirms that elongation occurs at the reducing end, because the mechanism for synthesis at this end (Fig. 2B) requires that the next sugar added displace the UDP attached to the previous sugar added. Thus, the [32P]UDP group is lost from a hyaluronyl-GlcNAc-[32P]-P-Uridine chain when the next sugar is added to give hyaluronyl-GlcNAc-GlcUA-P-Uridine. In the control situation, the next sugar addition again gives a radiolabeled product (i.e. hyaluronyl-GlcNAc-GlcUA-GlcNAc-[32P]-P-Uridine), but in the presence of excess unlabeled UDP-GlcNAc, incorporation of 32P into this latter product is greatly reduced (Fig. 5).
D. Hyaluronan chain elongation at the reducing end and nomenclature.
During chain elongation at the reducing end, the UDP-sugars are not the donors (as they are for additions that occur at the nonreducing end), but rather they are the acceptors. Therefore, the Class I hyaluronan synthase transferase activity that utilizes UDP-GlcNAc actually creates the GlcUA(β1,3)GlcNAc linkage. In contrast, the Class II pmHAS activity that utilizes UDP-GlcNAc creates the alternate GlcNAc(β1,4)GlcUA linkage. The multiple functions of a Class I hyaluronan synthase are summarized in Fig. 3, from the perspective of how the enzyme assembles hyaluronan at the reducing end. In each cycle of monosaccharide addition at the reducing end, the released UDP is derived from the previously added monosaccharide (Fig. 2B). The donor species transferred is the hyaluronan chain, which is still activated by its attachment to UDP.
The systematic name for such a transferase activity, according to IUBMB nomenclature, would specify the donor: acceptor, group transferred. Thus, the activity that adds a GlcUA-UDP residue to a GlcNAc at the reducing end of the growing hyaluronan chain is:
(HA)-GlcNAc-UDP: UDP-GlcUA, β(1,4)-hyaluronyl transferase.
Similarly, the activity that adds a GlcNAc-UDP to a HA-GlcUA-UDP chain is:
(HA)-GlcUA-UDP: UDP-GlcNAc, β(1,3)-hyaluronyl transferase.
E. Biological implications of synthesizing hyaluronan at the reducing end.
When chain elongation occurs at the reducing end, the hyaluronan product must have UDP, which is from the last sugar unit added, attached at the reducing end. When hydrolysis of the hyaluronan-UDP linkage occurs, then chain growth stops, because the hyaluronyl chain is no longer activated by its attachment to UDP and is not capable of serving as a donor in the transferase reaction. In contrast, for chain elongation at the nonreducing end, the original -UDP linkage at the reducing end does not participate in chain growth. If this linkage hydrolyzes, there is no effect on chain growth at the other end of the polymer.
Since the two HA-UDP linkages are less stable under physiological conditions than the two types of glycoside bonds in hyaluronan, it will be susceptible to hydrolysis, even at near-neutral pH. Therefore, hyaluronan chains in commercial preparations that have been processed in various ways will likely not still contain UDP at their reducing ends. However, it is intriguing to postulate that the presence of a novel HA-UDP structural element at the reducing end of a newly released hyaluronan chain could provide a specific recognition site for a class of proteins designed to recognize this linkage. Although the implications of such interactions could be physiologically important, they have not yet been explored.
In collaboration with Dr. Ellis Kempner, we used radiation inactivation analysis to determine whether active streptococcal hyaluronan synthases are a monomer or a larger oligomer14. We analyzed membranes prepared from Streptococcus cells expressing native spHAS or seHAS, as well as membranes made from E. coli cells expressing the recombinant enzymes. The results from these four experiments demonstrated that active native or recombinant hyaluronan synthases from both streptococcal species contain a single hyaluronan synthase protein, but the HAS protein is associated with an additional mass of about 23 kDa. The same result was later obtained for the Xenopus HAS1 enzyme, although the nature of the additional mass in this case was not determined15. In our study, we were fortunate in being able to identify this extra mass as cardiolipin. The active Class I hyaluronan synthase enzymes in membranes are, therefore, protein monomers in complex with other components; in the case of seHAS or spHAS, this active complex is a HAS protein associated with ~16 cardiolipin molecules.
The model in Fig. 3, in which the hyaluronan synthase itself is involved in hyaluronan transfer (translocation) across the membrane, is based on a variety of biochemical and genetic data1-3. These studies showed that the streptococcal hyaluronan synthase is the only protein required for hyaluronan synthesis in vitro. Also, the hasA gene encoding the synthase is the only gene required to transform a bacterial species that does not normally make HA, such as Bacillus subtilis and Enterococcus faecalis, into a strain that makes HA. If the UDP-sugar precursors are present in such cells, then bacteria transformed with the hasA gene are able to make HA, using cytoplasmic precursors, and transfer it to the cell exterior as an hyaluronan capsule or a secreted product that accumulates in the medium. Based on these and other data, there is no indication that a polysaccharide transporter is required in order for hyaluronan to appear outside the cell.
A. Overview.
When the original Chapter #5 was posted, no one had yet reported the detergent solubilization and successful purification of a bacterial or eukaryotic hyaluronan synthase. Hyaluronan synthases from any source have been extremely difficult to detergent-solubilize, with retention of activity, and then purify. Since then, however, one eukaryotic and three bacterial hyaluronan synthases have been purified5-7,16,17. Expression of the Group A and Group C streptococcal hyaluronan synthases in E. coli finally allowed us to purify these enzymes, 64 years after the discovery of hyaluronan. The recombinant streptococcal enzymes have been the easiest Class I hyaluronan synthases to purify and, therefore, they have been studied most extensively. The Class II P. multocida hyaluronan synthase was purified6,7 as a recombinant, truncated and soluble form of the protein expressed in E coli. Finally, the recombinant mouse HAS1 enzyme was purified and characterized kinetically5,17. No other eukaryotic hyaluronan synthases have been purified. The streptococcal hyaluronan synthases were the first enzymes that make a hetero-polysaccharide polymer to be studied in a purified state.
B. Hyaluronan synthase is the only protein required for hyaluronan biosynthesis.
The finding that hyaluronan synthesis requires only a single gene product was initially surprising, since the hyaluronan biosynthesis process is multifunctional (Fig. 3) and complex. Many investigators, for example, expected that at least two enzymes would be involved in order to catalyze the two glycosyltransferase reactions. We expected the synthase to be larger than ~49 kDa in order to perform the 6-7 functions shown in Fig. 3. Nonetheless, based on the results1-3 from genetic, biochemical, and radiation inactivation studies, and studies with the purified enzymes, it is clear that hyaluronan synthase is the only protein necessary for hyaluronan biosynthesis —when the UDP-sugars are provided. There is no evidence that hyaluronan synthases require a primer or any other proteins in order to synthesize HA.
C. Characteristics of the hyaluronan synthase proteins.
The three streptococcal hyaluronan synthase proteins are ~70% identical. Amino acid sequence identities among the eukaryotic hyaluronan synthases range from ~50% to 95%. The streptococcal proteins are ~25% identical to the eukaryotic proteins, which are about 35% larger. Except for the pmHAS enzyme, all the hyaluronan synthase proteins characterized to date share numerous amino acid motifs and are membrane proteins with multiple predicted transmembrane domains1-3. The hyaluronan synthase proteins appear to be very compact and tightly folded. Even during electrophoresis in the presence of the strong detergent sodium dodecyl sulfate, the HAS proteins behave as though they are ~15% smaller than their true size. Reducing agents do not change this behavior, which indicates that disulfide bonds are probably not present. The lack of disulfide bonds was confirmed biochemically for the streptococcal enzymes18-19, which have only four or six Cys residues. These latter studies also demonstrated that Cys residues are not required for hyaluronan synthase activity. The Group A and Group C streptococcal Cys-null hyaluronan synthases retain ~20% to 60% of the wild-type enzyme activity. Similar studies have not yet been performed with any of the eukaryotic hyaluronan synthases, in part because these larger enzymes have about 3-times as many Cys residues, making similar approaches more technically difficult.
D. Kinetic characteristics of hyaluronan synthases.
Although there are differences, the kinetic behaviors of hyaluronan synthases in membranes from various sources are generally very similar. The Km values for UDP-GlcUA utilization are typically lower, e.g. 30-75 μM for the streptococcal and human hyaluronan synthases, than for utilization of UDP-GlcNAc, e.g. 150-1000 μM for spHAS and the three human enzymes5,20. The less efficient utilization of UDP-GlcNAc by hyaluronan synthases may reflect the fact that this precursor sugar is used for the biosynthesis of many more important structures than is UDP-GlcUA, e.g. cell wall biosynthesis in bacteria or the core chitobiosyl linkage in N-linked oligosaccarides of eukaryotic glycoproteins. If the Km value for UDP-GlcNAc was lower, then hyaluronan synthesis might drain away this precursor so that cellular glycoconjugate biosynthesis was compromised. Consistent with this idea, we observed a sigmoidal Km profile for spHAS activity versus UDP-GlcNAc concentration20. This unexpected finding indicates that UDP-GlcNAc might be an allosteric regulator of the hyaluronan synthases. Although there is no in vitro kinetic evidence for similar behavior of seHAS or the eukaryotic hyaluronan synthases, it is possible that their in vivo behavior is different, especially regarding allosteric regulation by multiple small molecules.
It is more difficult to compare Vmax values for various hyaluronan synthases because different researchers use different methods to normalize the amount of hyaluronan synthesis to the amount of HAS protein. The hyaluronan polymerization rates for the streptococcal hyaluronan synthases in isolated membranes were estimated to be ~1200-2400 sugars/minute. At this elongation rate, it would take one active hyaluronan synthase molecule (or lipid complex) about 8-16 minutes to synthesize a single hyaluronan chain with a mass of 2 MDa. The rate of hyaluronan chain elongation in live cells has not been determined, but is likely to be faster than what has been measured in vitro.
When purified to homogeneity in the absence of exogenous phospholipids, the streptococcal hyaluronan synthases have very low activity16. When cardiolipin is added back, however, enzyme activity increases up to 10-fold (Fig. 6). The spHAS enzyme is very specifically activated by cardiolipin, whereas the seHAS enzyme is also activated by phosphatidylserine. Based on these results, other investigators have tried to use various phospholipids to help retain the activity of detergent-solubilized Class I eukaryotic hyaluronan synthases; unfortunately, these attempts have not yet been successful. The failure to purify a variety of these eukaryotic hyaluronan synthases, particularly the three human HAS isoforms, has been a disappointment and limitation in this field.
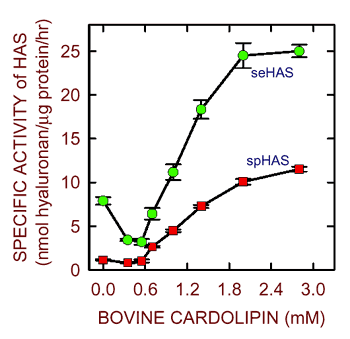
Fig. 6 Cardiolipins stimulate the activity of purified streptococcal synthases.
The purified recombinant seHAS (green) and spHAS (red) enzymes16 have low activity in the absence of exogenous lipid, but are stimulated by the addition of increasing amounts of a bovine cardiolipin mixture.
Nonetheless, we recently discovered that synthetic cardiolipins containing different fatty acids are profoundly different in their ability to activate purified seHAS (Fig. 7). For example, cardiolipin containing only C14 fatty acyl chains (myristic acid) is completely unable to activate seHAS, whereas cardiolipin containing only C18(Δ9) fatty acyl chains (oleic acid = palmitic acid with a double bond at carbons 9,10) activates the enzyme to a greater extent than routinely observed with commercial cardiolipin preparations from E. coli or bovine liver. Thus, within the Class I hyaluronan synthase family, there may be differences in the particular phospholipids (and specific fatty acids) required for activity, but it is possible that all these hyaluronan synthases require one or more particular type of phospholipid in order to synthesize hyaluronan. We have proposed that phospholipid molecules help the monomeric protein create a pore-like passage, through which the growing hyaluronan chain passes16. For example, the lipid portion of cardiolipin could interact with the lipid bilayer and hydrophobic patches of the hyaluronan chain, while the acidic head groups of cardiolipin molecules interact with the enzyme and hydrophilic portions of hyaluronan.
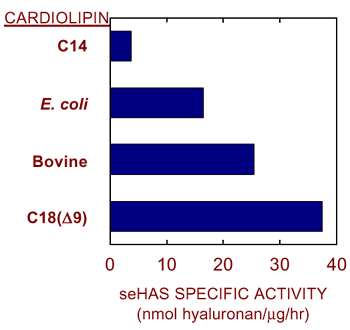
Fig. 7 Stimulation of streptococcal hyaluronan synthase depends on the type of cardiolipin.
Purified recombinant seHAS shows variable activity in the presence of 2 mM cardiolipin prepared synthetically (containing C14 or C18[Δ9] fatty acids; obtained from Avanti) or by extraction from bovine liver or E. coli membranes.
One of the major unanswered questions about hyaluronan synthase function in 1998 was how to reconcile the finding that only a single protein, in complex with phospholipids, is able to synthesize hyaluronan and also appears able to move the newly made polysaccharide across the cell membrane to the cell exterior. We have developed the Pendulum Hypothesis to explain how the enzyme might achieve the coordinated synthesis and movement of hyaluronan. The Pendulum Hypothesis, which has multiple variations, will be described in detail elsewhere, but the idea is conveyed by the animation presented in the following link (click here). In the first part, the growing hyaluronan chain is shown, for clarity, emerging in front of the synthase; in the second part, the last few sugar additions are repeated showing the hyaluronan chain passing through a pore within the hyaluronan synthsae protein-lipid complex.
The key features of the Pendulum Hypothesis, which can explain how Class I hyaluronan synthases work, are:
Some of the questions about the streptococcal hyaluronan synthases that remained in 1998 have since been answered, including that they build hyaluronan from the reducing end, and do not require a primer to initiate hyaluronan synthesis. There is also still no evidence that covalent enzyme intermediates are produced during hyaluronan synthesis. Despite this progress, at least three fundamental questions remain unanswered about how the Class I synthases function.
A. How many UDP-sugar binding sites are there in a HAS protein?
One of the major remaining questions is whether there are two different UDP-sugar binding sites or one site with alternating specificity. Although this latter model is unusual, it deserves consideration, in part, because the Class I hyaluronan synthases have only one classical “DXD” motif, which is a critical part of the nucleotide-sugar binding site of most glycosyltransferases. Unless a related region in these enzymes serves a similar function within a second nucleotide-sugar binding site, the single site model appears to be a likely alternative. In this case, one general binding site would alternate its sugar-binding specificity so that the two substrates are recognized in an alternating manner. Such a novel mechanism would require substantial conformational changes, and perhaps substantial movement within the enzyme.
B. How many glycosyltransferase active sites are there in a HAS protein?
A related remaining question is whether there are two glycosyltransferase active sites (e.g. in separate domains of the enzyme, such as for the Class II enzyme) or one versatile site, whose specificity for acceptor and donor changes in an alternating manner to create the two sugar linkages.
C. How is hyaluronan product size controlled?
We now recognize that the size of hyaluronan may profoundly affect its interactions with cells and its biological activities. Despite the great interest in how and why different hyaluronan synthases make hyaluronan products of different size distributions, we know very little about the enzyme properties that control hyaluronan chain length. We and others have demonstrated that specific hyaluronan synthase mutations can create variants that produce hyaluronan of altered size, but the mechanisms controlling chain length are not known.
Clearly, answering these and other remaining questions is critical if we want to understand the molecular details of how the Class I streptococcal and eukaryotic hyaluronan synthases synthesize the hyaluronan polymer. Hopefully, some of these questions will be answered and provide a reason to update this chapter again in the future.
I have described research results during the last five years that begin to answer some of the basic and important questions at the molecular level about how hyaluronan synthesis occurs. In 1993 we reported the first cloning of a glycosaminoglycan synthase. As of 2004, >20 hyaluronan synthases have been identified and cloned, and many have been expressed as recombinant proteins. Studies of the streptococcal hyaluronan synthases, the smallest members of the large Class I family, continue to provide the best opportunity to understand the details of hyaluronan biosynthesis at the molecular level.
Acknowledgments Research from my laboratory described in this article was supported by National Institutes of Health grant GM35978 from the Institute of General Medical Sciences.