
Manabu Igarashi
Department of Global Epidemiology, Hokkaido University Research Center for Zoonosis Control
Manabu Igarashi graduated from the Faculty of Engineering, Hokkaido University, in 1997. He completed his master’s degree at the Graduate School of Engineering, Hokkaido University in 2000, and obtained his Ph.D in 2004 at the Graduate School of Medicine, Hokkaido University. Since 2004 to the present he has been a postdoctoral fellow at the same university.

氏名:Kimihito Ito
Department of Global Epidemiology, Hokkaido University Research Center for Zoonosis Control
Kimihito Ito graduated from the Faculty of Engineering, Hokkaido University, in 1992.
He received his master's degree in electrical engineering in 1994, and earned his Ph.D. in the same field at the Graduate School of Engineering, Hokkaido University in 1999.
He was a postdoctoral fellow at the Graduate School of Engineering, Hokkaido University from 1999 to 2004, and was then appointed instructor at the Graduate School of Information Science and Technology, Hokkaido University.
He has been an associate professor in the Department of Global Epidemiology, Hokkaido University Research Center for Zoonosis Control since 2005.

Ayato Takada
Department of Global Epidemiology, Hokkaido University Research Center for Zoonosis Control
Ayato Takada graduated from the Faculty of Veterinary Medicine, Hokkaido University, in 1993, and obtained his Ph.D. degree in 1996 at the Graduate School of Veterinary Medicine, Hokkaido University. He worked as a postdoctoral fellow from 1996 to 1997, and appointed assistant professor at the Graduate School of Veterinary Medicine, Hokkaido University in 1997. In 2000, he moved to the Institute of Medical Science, University of Tokyo, as an assistant professor. Since 2005, he has been a professor in the Department of Global Epidemiology, Hokkaido University Research Center for Zoonosis Control.
For many viruses such as Hendra virus, severe acute respiratory syndrome-associated coronavirus, influenza virus, human immunodeficiency virus, hepatitis virus, and West Nile virus, the carbohydrate chains on their glycoproteins are responsible for the biological functions and thus play crucial roles in their life cycles (e.g., entry into host cells, proteolytic processing, and protein trafficking)1. Furthermore, addition or deletion of carbohydrate chains often results in antigenic changes of the viral glycoproteins. It has been suggested for influenza and human immunodeficiency viruses that the expression of carbohydrate chains on their glycoproteins facilitates viral escape from neutralization by antibodies1. In this section, we describe the N-linked glycosylation associated with the antigenic changes of the influenza virus glycoprotein hemagglutinin (HA).
It is known that influenza A viruses infect a variety of avian and mammalian species and the natural reservoirs of the viruses are wild aquatic birds belonging to the orders Anseriformes and Charadriiformes. Influenza A viruses are divided into subtypes based on antigenic differences of two virus surface glycoproteins, HA and neuraminidase (NA). Sixteen HA (H1-H16) and 9 NA (N1-N9) subtypes have been identified so far2,3. When some of these avian influenza viruses cross the species barrier by acquiring the ability to efficiently replicate in new host cells, these viruses may occasionally circulate in new host animal populations such as, pigs, horses, chickens, and humans. Acquisition of a new N-glycosylation site associated with the antigenic change of HA has been seen after introduction of viruses from the natural reservoir into these new host animals.
In the last century, influenza A viruses of only three subtypes, H1N1, H2N2, and H3N2, caused pandemics in humans4,5. Continuous antigenic changes of these influenza viruses occurred mainly in HA molecules. HA is a virus spike glycoprotein composed of two structurally distinct regions, the globular head and stem regions6 (Fig. 1). The globular head region is the major target site of antibodies that neutralize viral infectivity. The accumulation of a series of amino acid substitutions in this region under the selection pressure of host immune responses results in antigenic changes of HA that are sometimes associated with the acquisition of carbohydrate side chains. Because the carbohydrate side chains in the vicinity of antigenic sites mask the neutralizing epitopes on the HA surface, mutations associated with acquisition of carbohydrate chains are believed to efficiently generate HA antigenic variants.
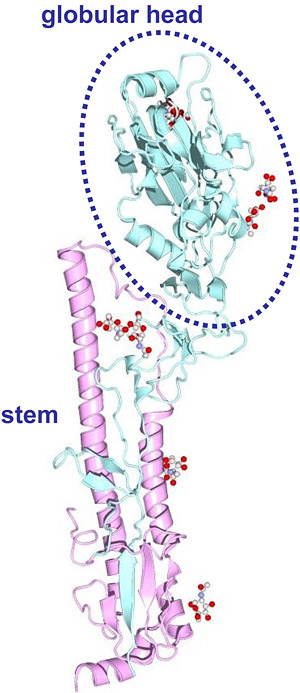
Fig. 1 Three-dimensional structure of HA monomer (PDB code: 1MQL).
We defined the globular head region as amino acid positions 52 to 277 (H3 numbering), for all HA subtypes.
It is well known that the number of N-glycosylation sequons (Asn-Xaa-Ser/Thr, where Xaa is any amino acid except Pro) in HA, especially on the globular head region, changes during circulation of the viruses in the human population (Fig. 2). The number of N-glycosylation sequons in the globular head region of H3 HA has been increasing during circulation in the human population for 39 years7-9. Most of the currently circulating H3N2 viruses have six or seven N-glycosylation sequons in the HA globular head region, whereas HA of the prototype H3N2 strain isolated in 1968, A/Hong Kong/1/68 (H3N2), had only two sites (Fig. 2A). H1N1 virus has also been circulating in humans for 30 years since its reemergence in 1977. Recent H1N1 viruses possess more N-glycosylation sequons in their HA sequences than the pandemic H1N1 strain9, A/South Carolina/1/18 (H1N1), which was detected from a few victims of the 1918 influenza10 (Fig. 2B). By contrast, HAs of H2N2 viruses did not acquire a new N-glycosylation sequon in the globular head region during the period of H2N2 epidemics (1957-1968) (Fig. 2C).
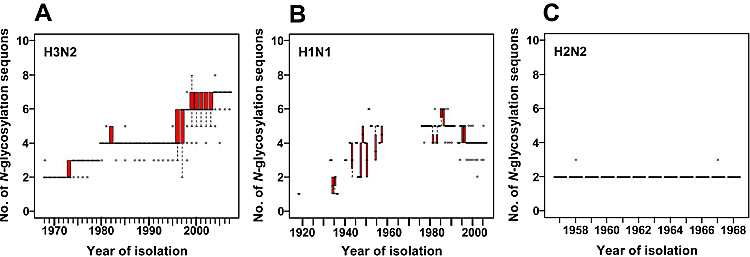
Fig. 2 Sequence data analyses of human influenza virus HAs.
The numbers of N-glycosylation sequons were examined for human H3N2 (A), H1N1 (B), and H2N2 (C) viruses. We used 950, 88, and 3415 sequences of H1N1, H2N2, and H3N2 human influenza viruses, respectively. In each box plot, the median value is indicated by a solid horizontal bar. The top and bottom edges of the box mark the first and third quartile, respectively. Outliers, which are shown as open circles, are cases with values more than 1.5 times the interquartile range away from the upper or lower quartile. The whiskers extending from the box indicate the highest and lowest values, excluding the outliers. The virus names and accession numbers used in this analysis are listed in a previous report12.
The epidemiological observations mentioned above have led us to hypothesize that the ability to acquire new carbohydrate side chains of HA was an important factor for sustained circulation of past pandemic influenza viruses (i.e. H1 and H3 subtypes) in the human population7,8,11. In this section, we describe their genetic backgrounds and structural locations for acquisition of N-glycosylation sequons of all 16 HA subtypes of avian influenza viruses, which may be potential of human pandemic viruses in the future.
We defined “N-glycosylation potential” as the number of potential candidate codons that were not sequons, but become sequons with 1-3 nucleotide mutations12 (Fig. 3). Sets of three codons that require single, double, or triple nucleotide substitutions to produce sequons are denoted Cand1, Cand2, and Cand3 sites, respectively.
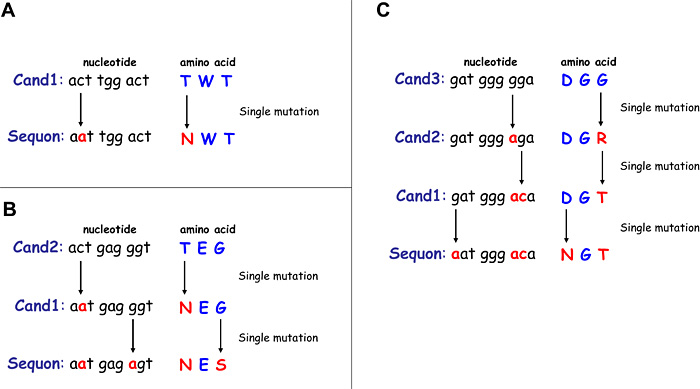
Fig. 3 Examples of potential candidate codons for N-glycosylation sequons.
Nucleotide sequences shown in (A) are representative codons of the Cand1 sites (i.e., Cand1 sites become sequons with single nucleotide substitution). (B) and (C) show examples of the Cand2 and Cand3 sites, respectively.
We focused on the virus strains of all the 16 subtypes isolated from the orders Anseriformes and Charadriiformes. We investigated the number of Cand1 and Cand2 sites in HA genes of these avian influenza viruses and compared their potentials to acquire N-glycosylation sequons. We found that the distribution of the number of Cand1 and Cand2 sites varied widely among the HA subtypes (Fig. 4B and 4C), while existing N-glycosylation sequons were similarly present in most of the HA subtypes (Fig. 4A). Therefore, we hypothesized that the ability of these avian viruses to rapidly acquire the N-glycosylation sequons associated with the antigenic change to evade antibody-mediated immune pressure in the human population might be different among the HA subtypes.
As shown in Fig. 4B, avian H1 and H3 viruses had more Cand1 sites in the globular head regions of HAs than avian H2 viruses. The same was equally true of the prototype strains of human influenza viruses in previous pandemics, A/South Carolina/1/18 (H1N1), A/Japan/305/57 (H2N2), and A/Hong Kong/1/68 (H3N2) (data not shown). As mentioned above, human H3N2 and H1N1 (Fig. 2A and 2B) viruses acquired additional N-glycosylation sequons in the HA globular head regions during their circulation, but human H2N2 viruses did not (Fig. 2C). Although several factors seem to influence the observation that the number of N-glycosylation sequons in the globular head region of H2 HA stayed constant, our analysis suggests that the H2N2 virus have already had an inherent disadvantage at the time of its appearance in the human population.
Fig. 4 Comparison of the N-linked glycosylation capacities among 16 HA subtypes of avian influenza viruses.
The number of N-glycosylation sequons (A), Cand1 (B), and Cand2 (C) sites of 16 HA subtypes of avian influenza viruses were compared by a two-sided nonparametric test. The p-values are listed in a previous report12. We used 47 (H1), 70 (H2), 104 (H3), 113 (H4), 747 (H5), 177 (H6), 66 (H7), 10 (H8), 89 (H9), 25 (H10), 52 (H11), 21 (H12), 15 (H13), 2 (H14), 5 (H15), and 6 (H16) sequences for comparison among avian viruses. Box plots are drawn as described in the legend of Fig. 2.
Avian influenza viruses of the H5, H7, and H9 subtypes have been circulating in domestic poultry. Since the first human case of H5N1 virus was identified in Hong Kong in 1997, direct avian-to-human transmission of several avian viruses, including H5N1, H7N7, H7N3, and H9N2, has been reported 4,13-15. These avian viruses pose a potential pandemic threat to humans. Our analyses demonstrated that avian H5 and H9 viruses had significantly larger numbers of Cand1 sites in the HA globular head regions than H2 viruses, and they were comparable to or had rather more such sites than H1 and H3 viruses (Fig. 4B). In addition, the number of Cand2 sites in HAs of avian H5 viruses was larger than that found in H1 HAs and comparable with that in H3 HAs (Fig. 4C). Avian H9 virus also had a number of Cand2 sites comparable to that of the H1 virus. Accordingly, most of the recently circulating avian H5N1 virus have already acquired a carbohydrate chain at position 158 (H3 numbering)16. Thus, we suggest that H5 and H9 viruses may have greater abilities to rapidly acquire N-glycosylation sequons than past pandemic viruses if introduced as new pandemic viruses in the human population. On the other hand, avian H7 virus had a significantly smaller number of both Cand1 and Cand2 sites than avian H2 virus, suggesting that avian H7 virus might have less ability to acquire N-glycosylation sites on the HA molecule surface.
We further investigated the spatial locations of Cand1 sites on 3D structures of HA molecules (H1, H2, H3, H5, H7, and H9 subtypes) (Fig. 5). Consistent with the differences among these HA subtypes in the numbers of Cand1 sites found in their original amino acid sequences (Fig. 4B), HA of the prototype strain of human H2N2 pandemic influenza virus expressed a smaller number of amino acids involved in N-glycosylation sequons and Cand1 sites on the solvent-accessible surface area of the HA molecule than H3, H5, and H9 HAs. Avian H5 and H9 viruses had larger numbers of Cand1 sites on the surface of the HA molecule than prototype strains of human H1N1, H2N2, and H3N2 pandemic viruses. Avian H7 virus had the smallest number of Cand1 sites on the HA surface. Thus, we confirmed that the differences among H1, H2, H3, H5, H7, and H9 subtypes in the number of Cand1 sites exposed on the HA molecule surface showed a tendency similar to the data for the primary sequence analysis (Fig. 4B).
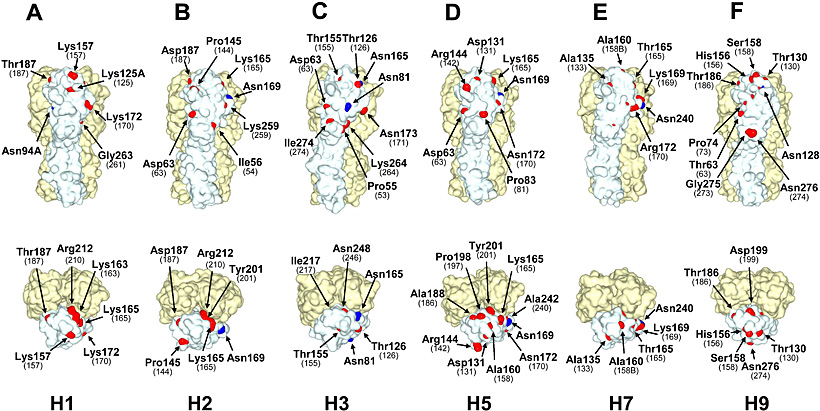
Fig. 5 The N-glycosylation sequons and Cand1 sites in solvent-accessible surface area representations of HA trimer structures.
Three-dimensional models of H1 (A/South Carolina/1/18) (A), H3 (STRAIN X-31) (C), H5 (A/duck/Singapore/3/97) (D), and H7 (A/turkey/Italy/02) (E) HAs were constructed from the coordinates obtained from the Protein Data Bank (PDB code: 1RUZ, 1HGF, 1JSM, and 1TI8, respectively). The structures of H2 (A/Japan/305/57) HA (B) and H9 (A/duck/Hokkaido/9/99) HA (F) were constructed by homology modeling. Residues shown in blue and red represent Asn at first positions in the present N-glycosylation sequons and amino acids that require nucleotide substitutions to produce sequons in Cand1 sites, respectively. Images were prepared by using MolFeat software (version 3.0, FiatLux Co.). Residue numbering is thoroughly on the basis of the H3 HA sequence18. Numbers in parentheses show the positions of Asn residues that may be linked to carbohydrate chains if respective Cand1 sites are mutated to N-glycosylation sequons.
Finally, to trace the history of the codon changes that resulted in the acquisition of sequons in the globular head region of HA of the human H3N2 and H1N1 viruses, we investigated Cand1, Cand2, and Cand3 sites during their epidemics, and confirmed whether these acquired sequons were derived from Cand1, Cand2, or Cand3 sites.
We found that all the additional sequons in H3 HAs were derived from Cand1, Cand2, or Cand3 sites (Fig. 6). The N-glycosylation sequons acquired in the first 20 years (Asn residues 63, 126, and 246) were Cand1 sites in HA of the prototype virus strain isolated in 1968. Two N-glycosylation sequons at Asn 122 and Asn 133 acquired in 1997 and an N-glycosylation sequon at Asn 144 acquired in 2003 were Cand2 and Cand3 sites, respectively, in HA of the prototype strain.
Fig. 6 Summary of the history of N-glycosylation sequons in HA of human H3N2 viruses isolated between 1968 and 2007.
Data represent the majority sequence obtained at each time point. The sequon at Asn residue 165 has been present for 39 years, and the sequon at Asn residue 81 has been lost since 1974.
We also traced the history of the codon changes of the human H1N1 viruses (Fig. 7). We found that the prototype strain, A/South Carolina/1/18, had an N-glycosylation sequon only at Asn residue 94A (H3 numbering) in the globular head region of HA, and the H1N1 virus that reemerged in 1977 possessed five N-glycosylation sequons at Asn residues 94A, 131, 158, 163, and 271. In addition, most recent H1N1 viruses had four N-glycosylation sequons at Asn residues 63, 94A, 129, and 163. The newly added N-glycosylation sequons were derived from the Cand1, Cand2, or Cand3 sites in HA of the prototype virus strain, suggesting the importance of these sites for the evolution of the H1N1 influenza virus in the human population.
Thus, our retrospective analyses of human H3N2 and H1N1 influenza viruses emphasized that the presence of Cand1 and Cand2 sites in HA genes of these pandemic strains was one of the key factors for the viruses to rapidly acquire N-glycosylation sequons during their evolutionary process in the human population.
Fig. 7 Summary of the history of N-glycosylation sequons in HA of human H1N1 viruses isolated between 1918 and 2006.
Data represent the majority sequence obtained at each time point. Because there is little information on the HA sequences of the human H1N1 virus isolated before its reemergence in 1977, the figure shows mosaic patterns.
In the present study, we analyzed a large number of nucleotide sequences of influenza virus HAs of different subtypes and compared their implicit potentials to acquire oligosaccharide chains. Importantly, there was a significant difference among HA subtypes in the genomic sequences needed to produce new N-glycosylation sequons, leading to the hypothesis that avian influenza viruses maintained in natural reservoirs have different abilities to rapidly evolve with N-glycosylation of HA if introduced into new host animals and exposed to immune pressure, and this depends on the HA subtype.
This hypothesis still needs to be supported by considering the statistical data on the structural environment for oligosaccharide attachment17, the probability data for nucleotide substitution, and so on. However, our approach may provide a possible way to predict the potential for sustained circulation of a hypothetical new pandemic influenza virus in the human population.