 |
More than 160 human glycosyltransferases have been cloned to date.
The number of cloned enzymes is increasing and may reach 200 within
one year (Narimatsu, Glycoconj. J. 2004). Glycosyltransferases catalyze
the transfer of sugar residues from nucleotide sugars (donor) to a growing
carbohydrate chain (acceptor). Glycosyltransferases can be grouped into
functional families based on their sequence similarities, which reflect
their enzymatic characteristics: donor specificity, acceptor specificity,
and linkage specificity between donor and acceptor. By the accumulation
of glycosyltransferase gene data, it becomes evident that glycosyltransferases
have strict specificity of linkage between donor and acceptor as well
as donor specificity, whereas they show much lower specificity for acceptor.
Here the general line of evolutionary history of glycosyltransferase
gene families is discussed. The authors conducted molecular evolutionary
analysis on some glycosyltransferase genes and mainly describe glycosyltransferase
genes for N- or O-glycan synthesis (Kaneko, TIGG, 2000).
The phylogenetic trees showed that the glycosyltransferase genes increased
in number through gene duplication. We estimated the divergence time
of each branch root and suggest that the glycosyltransferase genes increased
in number through gene duplication and genome duplication (Fig.
1). The results show that gene duplication of the intra-gene
family is concentrated around the early period of vertebrate lineage.
Prior to gene duplication of the intra-gene family, gene duplication
of the inter-gene family seems to have occurred. Briefly, in the ancestral
genome, the ancestral glycosyltransferase may have multiplied by gene
duplication and acquired variable activities via accumulation of point
mutation, gene conversion, exon shuffling, and other phenomena. After
the ancestral genome was equipped with a basic set of ancestral glycosyltransferase
genes, genome duplication events may have occurred.
|
|
|
Recent investigations have demonstrated that two genome-duplication
events occurred (tetraploidization); the first is close to the origin
of the vertebrates and the second is close to the origin of the gnathostomes
(Kasahara, Trend in genetics, 1997). We noticed that glycosyltransferase
genes show ?homologous cluster,? which was defined as more than two
chromosomal homologous segments. For example, the MHC gene clusters
were detected on chromosomes 1, 6, 9, and 19. Genes of the Hox clusters
were also mapped to chromosomes 2, 7, 12, and 17. These four regions
may be the remnant of the four homologous segments, called homologous
cluster that arose from the genome tetraploidization.  1,3FucT
genes were placed in chromosomes 6, 9, and 19. The segment on chromosome
1 was missing. Some 1,3FucT
genes were placed in chromosomes 6, 9, and 19. The segment on chromosome
1 was missing. Some 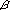 3GalT genes were placed in chromosomes 1,
6, and 19. The segment on chromosome 9 was missing. Mgat genes were
located on chromosomes 2, 7, 12 and 17. Some sialyltransferase genes
were located on chromosomes 1, 9, 2, 12, and 17. The period of gene
duplication estimated by molecular evolutionary analyses was compatible
with that of genome duplication. In conclusion, a basic set of ancestral
glycosyltransferase genes was fully present in the ancestral genome
before the genome duplication events. After the genome tetraploidization,
the increase in glycosyltransferase genes allows amino acid change and
permits the creation of a variety of specific activities of these enzymes.
Additionally, the information of the genome localization of glycosyltransferase
genes will help us to deduce the long evolutionary history of glycosyltransferases. 3GalT genes were placed in chromosomes 1,
6, and 19. The segment on chromosome 9 was missing. Mgat genes were
located on chromosomes 2, 7, 12 and 17. Some sialyltransferase genes
were located on chromosomes 1, 9, 2, 12, and 17. The period of gene
duplication estimated by molecular evolutionary analyses was compatible
with that of genome duplication. In conclusion, a basic set of ancestral
glycosyltransferase genes was fully present in the ancestral genome
before the genome duplication events. After the genome tetraploidization,
the increase in glycosyltransferase genes allows amino acid change and
permits the creation of a variety of specific activities of these enzymes.
Additionally, the information of the genome localization of glycosyltransferase
genes will help us to deduce the long evolutionary history of glycosyltransferases.
|
|
|
| References |
(1) |
Narimatsu H: Construction of a human glycogene library and comprehensive functional analysis. Glycocojugate J. 21, 17-24. 2004 |
|
(2) |
Kaneko M, Nishihara S, Narimatsu H, Saitou N: The Evolutionary
History of Glycosyltransferase Genes. Trends in Glycosciences and
Glycotechnology, 13, 147-155, 2001 |
|
(3) |
Kasahara M, Nakaya J, Satta Y, Takahata N: Chromosomal duplication
and the emergence of the adaptive immune system. Trend in Genetics,
13, 90-92, 1997 |
|
|



