
Dr. Joe G. Hollyfield received a Ph.D. in Zoology from the University of Texas in 1966. He then did Postdoctoral Fellowship training funded by Fight for Sight, Inc. (Schaumburg, IL) at the Hubrecht Laboratory in Utrecht, The Netherlands. He was appointed Assistant Professor in the Department of Anatomy assigned to Ophthalmology at Columbia University College of Physicians and Surgeons in New York City in 1969. In 1977, he moved to the Cullen Eye Institute, Baylor College of Medicine, where he was Director of the Research Center supported by The Foundation Fighting Blindness (Hunt Valley, MD). In 1995, Dr. Hollyfield joined The Cleveland Clinic Foundation as Director of Research in the newly established Cole Eye Institute. His research contributions to the developmental and cell biology of the retina and retinal pigment epithelium have been recognized by such honors as The Endre Balazs Prize from the International Society for Eye Research (San Francisco, CA); The Alcon Research Institute Award (Ft. Worth, TX); the Marjorie W. Margolin and the Sam and Bertha Brochstein Awards from the Retina Research Foundation (Houston, TX); awards from Research to Prevent Blindness (New York, NY); the Ocular Cell and Molecular Biology Prize from Allergan Laboratories (Irvine, CA); and the Distinguished Alumnus Award from Hendrix College (Conway, AR). Dr. Hollyfield has served as Secretary and President of the International Society for Eye Research and as a member of the Board of Trustees and President of the Association for Research in Vision and Ophthalmology (Bethesda, MD). Since 1992, Dr. Hollyfield has been Editor-in Chief of the journal Experimental Eye Research.
The inner and outer segments of rod and cone photoreceptors project from the outer surface of the retina toward the retinal pigment epithelium. A novel matrix, referred to as the interphotoreceptor matrix (IPM), surrounds the outward extension of these cells (Fig. 1). Unlike the matrix of connective tissues that reside below basal lamina and are derived from embryonic mesoderm and neural crest, the IPM is located in a compartment bordered by the apical surfaces of two epithelia that develop from neural ectoderm. The apical surfaces of these epithelia are placed in apposition following the collapse of the optic vesicle to form the double-walled optic cup (Fig. 2). The IPM has been implicated in several important activities required for photoreceptor function and maintenance. These include retinal adhesion, photoreceptor alignment, growth factor presentation, retinoid transport, and photoreceptor outer segment recognition for phagocytosis1-8.
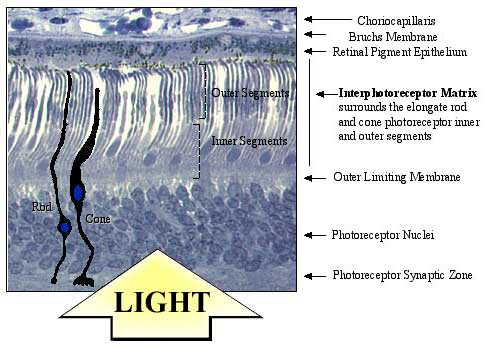
Fig. 1 Location of the interphotoreceptor matrix (IPM).
Photoreceptor cells that generate the primary neural signal in response to light are located in the outer lamina of the vertebrate retina. These primary sensory neurons are highly elongated with cellular organelles controlling specific photoreceptor functions compartmentalized into discrete regions. The light-sensitive outer segment that contains visual pigment is the most distal portion of the cell. It is connected by a ciliary derivative to the inner segment, where the rough endoplasmic reticulum, Golgi apparatus and mitochondria reside below the inner segment is the nucleus, located in the outer nuclear layer of the retina. A short axon terminates in a synaptic terminal, which abuts the outer plexiform layer. The interphotoreceptor matrix fills the compartment between the retinal pigment epithelium and the outer limiting membrane of the retina. Photoreceptor inner and outer segments projecting from the outer surface of the retina are surrounded by this matrix. All metabolite trafficking and exchange between the photoreceptor and retinal pigment epithelium are conducted through the interphotoreceptor matrix. (The micrograph shows normal human adult retina).
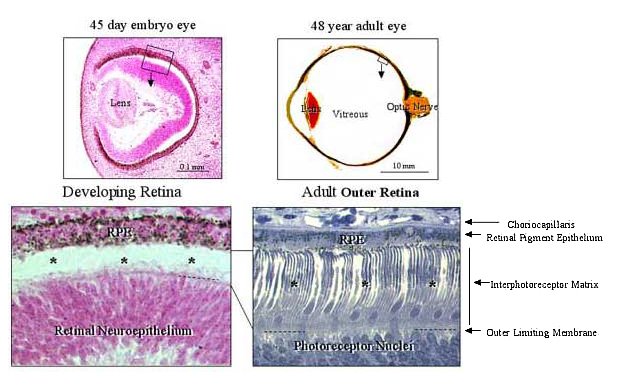
Fig. 2 Embryonic origin of the IPM.
The tissue interface where the IPM is located emerges during early eye development, following the collapse of the optic vesicle to form the double-walled optic cup. This early morphogenetic event brings together the apical surfaces of the inner (the future retina) and outer (the future retinal pigment epithelium, RPE) neuroepithelium of the optic cup wall. This illustration compares the outer eye wall from a 45-day post-fertilization human embryo before the development of photoreceptor cells (*) with an equivalent region from a 48-year old adult eye in which the elongated photoreceptor cells (*) extend from the outer retinal surface. The IPM present between these epithelial-derived layers is equivalent to a glycocalyx present along the apical surface of an epithelium, in contrast to a connective tissue matrix located below a basal lamina.
This chapter will present a brief history of the analysis of hyaluronan in the IPM and a discussion of novel hyaladherins recently identified in this matrix. It will conclude with a model for the organization of IPM. This model places hyaluronan in a key role as the scaffold on which this matrix is organized.
Early biochemical studies of the IPM attempted to rinse extracellular molecules from the outer retina for subsequent analysis. Small amounts of hyaluronan and chondroitin sulfate were identified using this approach1-8. It was not until the lectin signatures of novel molecules present in this matrix emerged and more aggressive IPM extraction procedures were utilized that an appreciation for the resistance of hyaluronan and its associated molecules to aqueous extraction was recognized9,10. In studies using bovine11 and human retina 12, the hydrophylic nature of the IPM was first appreciated. When retinas are placed in distilled water, this extracellular compartment rapidly swells and detaches from the outer retina, providing a rapid procedure for IPM isolation (Fig. 3).
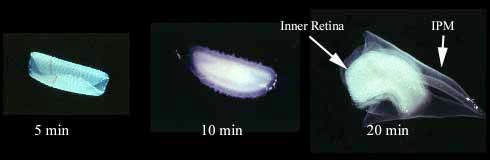
Fig. 3 Isolation of the IPM.
The highly hydrophylic nature of the IPM causes it to swell to over 2.5 times its normal dimension when retinas or retinal fragments are placed in distilled water. This composite illustration documents the progression of detachment of the IPM from a 3-mm disc of retina when placed in distilled water. The greatly expanded diaphanous and transparent sheet (right) contains the IPM. This material can be easily separated from the chalk-like inner retina for morphological or biochemical analysis.
The isolated IPM was used to document the distribution of different lectin-binding domains in the rod and cone matrix compartments, which had been previously described using conventional microscopic procedures13,14. These studies demonstrate that the lectin peanut agglutinin (PNA) decorates primarily the cone matrix compartments, whereas wheat germ agglutinin (WGA) and Maackia amurensus agglutinin (MAA) show higher affinity for the rod-associated matrix11,15,16 (Fig. 4). These differences in lectin staining of the rod and cone matrix have been documented in the isolated IPM in a variety of mammalian species (Fig. 5). Furthermore, ultrastructural analyses of these isolates clearly demonstrate the extracellular nature of this matrix as well as the aqueous stability of this complex (Fig. 4).
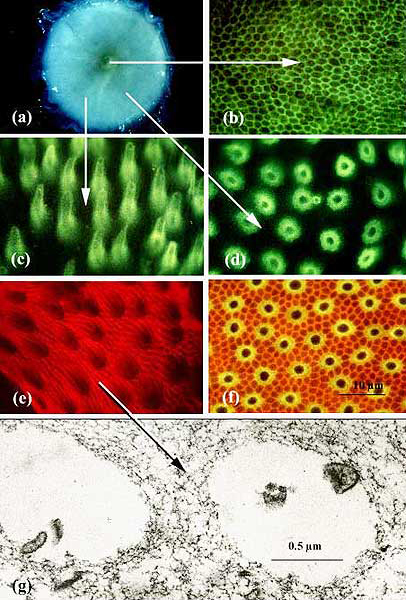
Fig. 4 Distinct lectin-binding domains in the IPM.
The image (a) is a 3-mm disc of the adult human retina which includes the fovea (yellow center) in partial stages of detachment of the IPM with distilled water treatment. When the IPM sample is detached and stained with the agglutinin PNA conjugated to FITC, green fluorescence is present as a continuous reticulum in the matrix surrounding foveal cones (b). In more peripheral locations, where cone density is lower, the cone-associated matrix also binds PNA, but each matrix domain is much more expansive than that surrounding individual foveal cones (c, d). The PNA-decorated cone matrix domains are viewed from the side (c). These bright fluorescent domains would occupy the apparent transparent areas surrounding cone inner and outer segments presented in Fig. 1. When viewed directly en face, (d), there is near complete fidelity in PNA staining of the cone matrix compartments, with the exception of an occasional rod matrix compartment. Wheat germ agglutinin (WGA) conjugated to rhodamine only weakly decorates the cone matrix compartments, but intensely stains the rod matrix (e). When PNA-FITC and WGA-rhodamine are used simultaneously, the colocalization of both lectins surrounding cones is apparent from the yellow color in the cone matrix compartments (f). Electron microscopy of the isolated IPM provides proof that the distilled water IPM isolate indeed represents the matrix components surrounding photoreceptors and not the cellular components (g). The IPM in this image consists of a fine reticulum of interconnected fibrillar-like processes surrounding the electron lucent openings previously occupied by rod photoreceptors (some membrane debris is present in these luminal areas). The IPM isolate was stained en bloc with WGA-ferritin, and ferritin densities are apparent associated with the fibrillar profiles comprising the matrix.
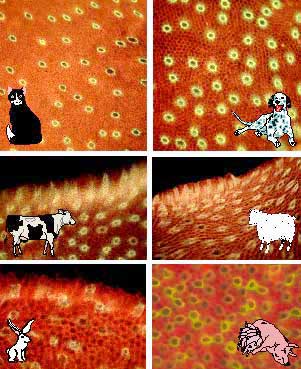
Fig. 5 Lectin binding domains in the IPM of other mammalian retinas.
The IPM in a variety of mammalian species can be isolated by methods similar to that demonstrated in Fig. 3, above. In all mammals thus far studied, similar lectin staining patterns as those described in Fig. 4 for the human IPM are evident.
The lectin-binding characteristics of rod- and cone-associated domains in IPM isolates were useful in identifying and characterizing specific molecules in this compartment. When isolated matrix molecules were displayed with SDS/PAGE and blotted, one major PNA stained band was evident at 150-kD (Fig. 6).
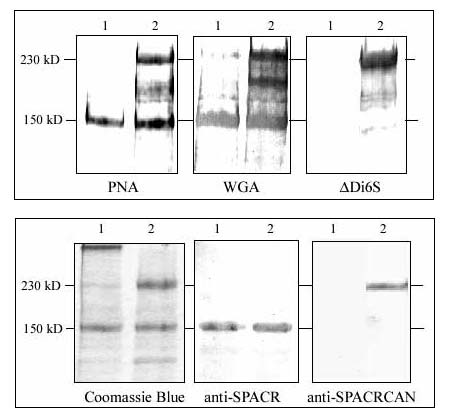
Fig. 6 Identity of the IPM molecules.
Lectin and western blots of IPM proteins and proteoglycans separated with SDS/PAGE. Paired sample lanes are presented for each lectin or antibody blot. Lane 1 contains the undigested IPM sample, and Lane 2 contains an IPM sample digested with chondroitinase ABC. Upper panel: PNA decorates only a single band at 150-kD in the undigested sample. Following digestion, several higher molecular weight bands are present with a prominent band at 230-kD. Although many more bands are evident in the WGA blot, a prominent band is present before and after digestion at 150-kD, whereas the 230-kD band is most pronounced after chondroitinase digestion. The ΔDi6S monoclonal antibody that recognizes the chondroitin-6-sulfate disaccharide remaining following chondroitinase ABC digestion does not interact with any bands in the undigested sample but intensely labels the 230-kD band in the chondroitinase ABC digested sample. Although there is some background staining further down the lane, the 150-kD band is not labeled. Lower panel: The 150-kD band (SPACR) and the 230-kD band (SPACRCAN) were cut from polyacrylamide gels similar to that shown on the left, stained with Coomassie Blue. These isolated bands were used to prepare polyclonal antibodies. In the western blot, using the anti-SPACR antibody a single immunoreactive band is present at the 150-kD level, and the intensity of staining of this band is similar before and after chondroitinase ABC digestion. In contrast, with the anti-SPACRCAN antibody, immunoreactivity is present only after chondroitinase ABC digestion, and immunoreactivity is predominantly associated with the 230-kD SPACRCAN band.
Because of reports describing the presence of chondroitin sulfate proteoglycan in this compartment3,17,18, IPM samples were also digested with chondroitinase to free any chondroitin sulfate from the core proteins of proteoglycans before SDS/PAGE. At least two new bands that stained prominently with PNA were observed following chondroitinase treatment, indicating that some of the PNA binding molecules were chondroitin sulfate proteoglycans. Although WGA decorated many bands in lectin blots of these preparations, the 150-kD and 230-kD bands observed after chondroitinase treatment were prominent (Fig. 6). Polyclonal antibodies were prepared against the 150-kD and 230-kD bands cut from SDS/PAGE. In western blots these antibodies label the 150-kD and 230-kD bands, respectively (Fig. 6), and when used for immunocytochemistry, they show unique localization to the IPM in tissue sections (Fig. 7).
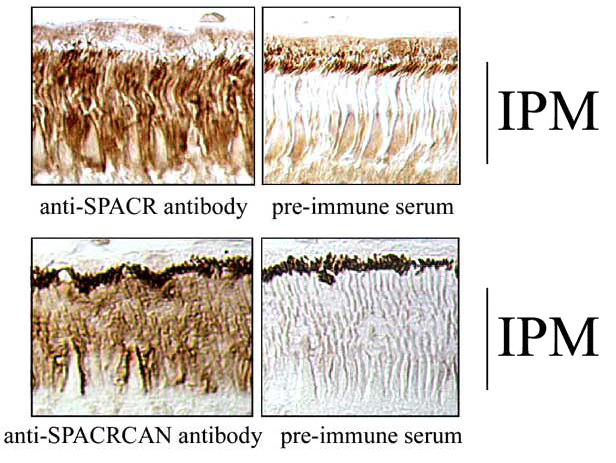
Fig. 7 Immunolocalization of SPACR and SPACRCAN in the IPM.
Sections of primate retina (Macaca mulatta) used for localization of immunoreactivity with the SPACR and SPACRCAN antibodies described in Fig. 6. Note the intense immunoreactivity over the IPM compartment with each antibody and the absence of IPM staining in the pre-immune serum controls.
We characterized the 150-kD glycoprotein and identified N-linked and an abundance of O-linked oligosaccharides, some of which were capped with sialic acid. Accordingly, we named this molecule SPACR (SialoglycoProtein Associated with Cones and Rods)15. The gene for SPACR was cloned and mapped to chromosome 6q14. The gene for the 230-kD proteoglycan was cloned and mapped to chromosome 3q11.2. Because of the high sequence homology to SPACR and the presence of covalently linked chondroitin sulfate, we named the 230-kD molecule SPACRCAN19 (Fig. 8).
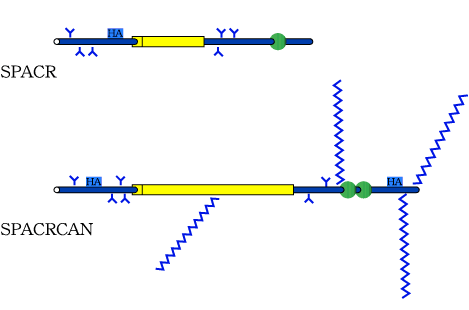
Fig. 8 Stick models of human SPACR and SPACRCAN showing the major motifs present in the deduced amino acid sequence.
Major homologies are shared by SPACR and SPACRCAN, as described in detail elsewhere19. A large mucin-like domain (yellow boxed area) in the central region of the polypeptide contain numerous potential sites for O-linked oligosaccharides. Clusters of N-linked oligosaccharides consensus sites (blue Y-shaped projections) are present on either side of the mucin-like domains. Both molecules contain epidermal growth factor (EGF)-like domains (green circles) near their C-termini: one in SPACR, two in SPACRCAN. Both molecules contain putative RHAMM-type hyaluronan-binding motifs (blue squares) on the N-terminal side of the mucin domain. SPACRCAN has a second hyaluronan-binding motif near the C-terminus. SPACRCAN contains four potential consensus sites for glycosaminoglycan chain attachment (chains associated with these sites are depicted with saw-tooth blue lines). Conventional mammalian consensus sites for glycosaminoglycan attachment are not present in SPACR, although an avian glycosaminoglycan attachment site is present on the N-terminal side of the mucin domain. Migratory mass changes in SDS/PAGE following enzymatic removal of chondroitin sulfate, and of N- and O-linked glycoconjugates demonstrate that carbohydrates account for two thirds of the 400-kD apparent molecular weight of SPACRCAN. Similar studies on the glycoprotein SPACR demonstrate that N- and O-linked glycoconjugates account for approximately one third of its migratory mass.
Using the hyaluronan-binding complex derived from aggrecan, we also identified hyaluronan in the distilled water isolate of the IPM19,20. In our analysis of the deduced sequence of SPACR and SPACRCAN polypeptide, we observed potential hyaluronan-binding motifs of the RHAMM type21, suggesting that these IPM molecules may interact with hyaluronan (Fig. 7)a. To assess directly the interaction of these molecules, we conducted a series of precipitation studies using exogenous hyaluronan, SPACR and chondroitinase digested SPACRCAN. These studies demonstrate that these novel IPM molecules pellet with hyaluronan when precipitated with cetylpyridinium chloride, but do not pellet when cetylpyridinium chloride is used in the absence of hyaluronan. This strongly suggests that these molecules bind directly to hyaluronan19,20.
a See article by Turley and Harrison in this series.
To define the distribution of hyaluronan in the IPM, we used the biotinylated hyaluronan-binding complex (bHABC) derived from aggrecan as a specific probe for hyaluronan on sections of human retina that were prepared for microscopy under conditions that retain hyaluronan (Fig. 9). Intense labeling was observed on the vitreal surface of the retina, and in the connective tissue of the choroid, with weaker staining present throughout the IPM. Streptomyces hyaluronidase has high specificity for hyaluronan; because of this specificity, many investigators routinely use this enzyme in digestions to establish the specificity of bHABC localization in tissue sections. Surprisingly, the hyaluronan staining in the IPM was resistant to Streptomyces hyaluronidase degradation. Because of the resistance of IPM hyaluronan to Streptomyces hyaluronidase digestion it was necessary to perform blocking controls by preincubating bHABP with hyaluronan oligomers approximately 22 sugar units long. These preadsorption controls completely eliminated bHABC staining of the IPM, indicating that the probe indeed interacts with hyaluronan in the IPM (Fig. 9).
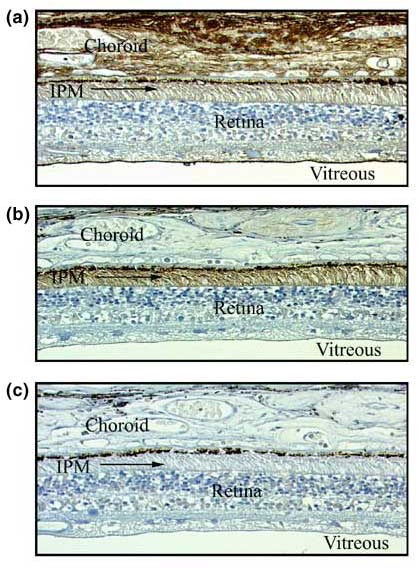
Fig. 9 Hyaluronan distribution in the IPM.
Sections were prepared from the outer eye wall of the human eye from a 54-year-old male donor. (a) This section was stained with bHABC to demonstrate the localization of hyaluronan. Intense staining is present in the choroid and on the surface of the retina that borders the vitreous. Moderate staining is present in the IPM. (b) This section was digested with Streptomyces hyaluronidase prior to staining with bHABC. Although this enzyme treatment eliminates staining in the choroid and on the vitreal surface of the retina, hyaluronan staining in the IPM is enhanced above the level present in the undigested sample. (c) When the bHABC probe is incubated with a 22-sugar fragment of hyaluronan prior to application to either undigested sections or to sections digested with Streptomyces hyaluronidase, all staining is abolished, and the only densities remaining are the dark melanin pigment in the retinal pigment epithelium (see also Fig. 10).
To explore this curious resistance of hyaluronan in the IPM to Streptomyces hyaluronidase digestion, we used other enzymes that degrade hyaluronan; Streptococcus hyaluronidase, testicular hyaluronidase and chondroitinase ABC in similar experiments. Each of these enzymes degrade hyaluronan as well as chondroitin sulfate, and each completely eliminated bHABC staining of the IPM. These results demonstrate that enzymes capable of degrading both chondroitin sulfate and hyaluronan are able to degrade hyaluronan in the IPM, whereas an enzyme capable of degrading only hyaluronan is unable to do so (Fig. 10).
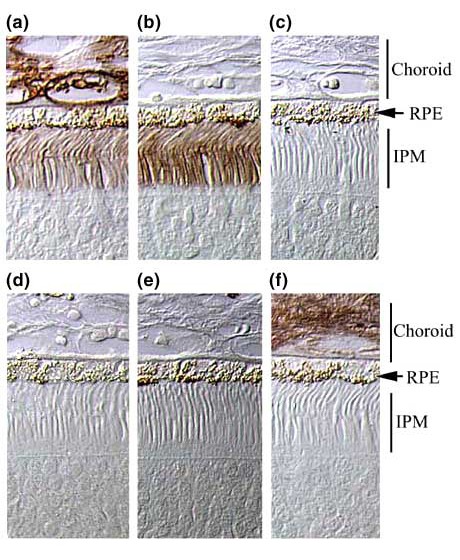
Fig. 10 Susceptibility of hyaluronan in the IPM to enzyme degradation.
Control incubations with bHABC show light staining of hyaluronan in the IPM and more intense staining of hyaluronan in the choroid (a). After digestion with Streptomyces hyaluronidase (which is highly specific for hyaluronan), hyaluronan staining in the choroid is eliminated, but hyaluronan staining in the IPM is enhanced over the level in the undigested control (b). When the bHABC probe is preincubated with 22-mer fragments of hyaluronan, all hyaluronan staining in the IPM of Streptomyces hyaluronidase-digested tissue is eliminated (c). When tissues are digested with Streptococcus hyaluronidase (which degrades hyaluronan and chondroitin sulfate), staining in the IPM and choroid is eliminated (d). When tissues are digested with testicular hyaluronidase (which degrades hyaluronan and chondroitin sulfate), staining in the IPM and choroid is eliminated (e). When tissues are digested with chondroitinase ABC (which degrades chondroitin sulfate and hyaluronan), staining in the IPM is eliminated, but staining in the choroid remains (f). These digestions indicate that enzymes capable of degrading both chondroitin and hyaluronan are able to degrade hyaluronan in the IPM, but the enzyme capable of degrading only hyaluronan is incapable of degrading IPM hyaluronan. We conclude from these results that the chondroitin sulfates covalently linked to the proteoglycan SPACRCAN in the IPM prevent Streptomyces hyaluronidase from gaining access to the hyaluronan scaffold. When the chondroitin sulfates are degraded, hyaluronan buried deeper in the complex is then vulnerable to degradation.
We conclude from these results that the chondroitin sulfate chains covalently linked to the proteoglycan SPACRCAN prevent Streptomyces hyaluronidase access to the hyaluronan scaffold in the IPM. When they are degraded, hyaluronan normally shielded by chondroitin sulfate becomes vulnerable to degradation. A model of the IPM showing the associations of SPACR, SPACRCAN and hyaluronan suggested by these results is presented in Fig. 11.
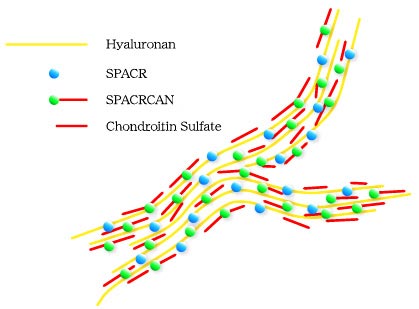
Fig. 11 Hyaluronan, SPACR and SPACRCAN associations in the IPM.
A model of the hyaluronan scaffold in the IPM with associated SPACR and SPACRCAN based on the data from enzyme degradation of hyaluronan presented in Fig. 10. The chondroitin sulfate chains associated with SPACRCAN protect the hyaluronan scaffold from hyaluronan-specific enzyme degradation. Enzymes that attack chondroitin sulfates and hyaluronan are capable of degrading hyaluronan because degradation of chondroitin sulfate chains exposes the deeper hyaluronan scaffold to the enzyme.
Analysis of the novel IPM glycoprotein SPACR, the proteoglycan SPACRCAN, and their demonstrated interactions led us to propose that hyaluronan may function in the IPM as the basic scaffold to which these and other IPM molecules are anchored. Other molecules that bind hyaluronan have been identified in the plasma membrane of cells that border the IPM (Müller cells, photoreceptor cells and retinal pigment epithelial cells). These include CD44, a well recognized hyaluronan-binding protein present in the apical microvillae of Müller cells6 and RHAMM, localized to the apical region of the RPE22. Neuroglycan C, a proteoglycan that is associated with the plasma membrane of photoreceptors, contains a RHAMM-type hyaluronan-binding motif in its extracellular domain23. A recently discovered gene expressed in photoreceptors and linked to the loci for retinitis pigmentosa-1 codes for oxygen-regulated photoreceptor protein-1 (ORP-1)24. The deduced sequence of the ORP-1 polypeptide contains 14 putative hyaluronan-binding motifs. Thus, the presence of hyaluronan-binding motifs on the cells that border the IPM provide the structural components through which these cells can interact with the hyaluronan scaffold. Attachment of the hyaluronan scaffold to cells on both sides of the IPM would allow the IPM to form the structural link between the retina and RPE by way of these protein-carbohydrate and carbohydrate-carbohydrate interactions (Fig. 12).
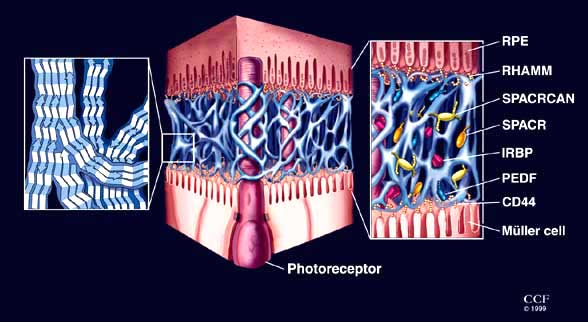
Fig. 12 Hyaluronan distribution in the IPM.
Cartoon showing the levels of organization of hyaluronan and hyaladherins associated with the basic IPM scaffold. The left panel shows the possible antiparallel alignment of linear hyaluronan molecules forming the basic matrix scaffold structureb. The center panel depicts the continuous three-dimensional scaffold complex (not to scale) in the extracellular compartment, adapted from electron microscope images of the IPM similar to that shown in the bottom panel of Fig. 4. The right panel depicts the interaction of the scaffold (not to scale) with hyaluronan-binding motifs on cells that border the IPM (CD44 is present in the apical microvillae of Müller cells, and RHAMM has been identified on apical RPE processes) and secreted molecules within the IPM (SPACR, SPACRCAN, pigment epithelium derived factor [PEDF] and interphotoreceptor matrix-binding protein [IRBP]). Since IRBP can be removed from the IPM with saline rinses, it is not considered part of this insoluble IPM complex. (Drawing by David Schumick, Medical Illustrator, The Cleveland Clinic Foundation.)
b See article by Scott in this series.
While the evidence reviewed in this chapter fits well with the suggestion that hyaluronan forms the basic scaffold for organization of the IPM, additional studies are needed to test several aspects of the hypothesis implicit in this new model of matrix organization. Important areas for further investigation are: (A) to determine the pattern and rates of addition and loss (turnover) of hyaluronan and other molecules present in the IPM; (B) to identify other molecules in the IPM that associate with the scaffold; (C) to define the cellular sources of hyaluronan present in the scaffold; (D) to identify other molecules that function to attach the scaffold to cells that border the IPM; (E) to define the changes in the scaffold that precede retinal detachment; and (F) to understand the processes that repair the scaffold following retinal reattachment. As these and other studies on the IPM are completed, a more complete understanding of the importance of hyaluronan in this novel matrix will be forthcoming.
Acknowledgments -We thank the Cleveland Eye Bank for the human tissues used in this study. The study was supported by grants from the National Institutes of Health (Bethesda, MD), The Foundation Fighting Blindness (Hunt Valley, MD), The Retina Research Foundation (Houston, TX), and funds from The Cleveland Clinic Foundation. Colleagues and collaborators who have been involved in portions of the work and discussions leading to the development of this organizational model of the IPM include: Hugh H. Varner, Peter M-T. Ho, Steven J. Fliesler, Akihiko Tawara, Masayuki Iwasaki, Robert A. Landers, Lifang Tien, Kathy Myers, Matthew M. LaVail, Victoria C. Foletta, Jung Wha Lee, Shreeta Acharya, Ignacio R. Rodriguez, W. Scott Young, III, Ronald J. Midura, Preenie deS. Senanayake, Raija Tammi, Markku Tammi, Anthony Calabro, Karen G. Shadrach, Qiuyun Chen, Kazutoshi Nishiyama and Vincent C. Hascall. I am particularly grateful to my wife, Mary E. Rayborn, for her many years of personal and professional support. Without her excellent technical expertise and persistence, many of the images included in this article would not have been generated.