
Paul L. DeAngelis has explored glycobiology throughout his career. As an undergraduate at Harvard (B.A. 1984), Dr. DeAngelis investigated the carbohydrate structures of polysaccharides from fungal pathogens that their plant hosts detected as "foreign" by creating and testing a series of synthetic structural mimics. As a graduate student at the University of California, Irvine (Ph.D. 1990), he elucidated some details surrounding fertilization, such as defining the critical elements of egg surface polysaccharides and the essential residues of a sperm adhesive protein. As a postdoctoral researcher (1990-93) in the laboratory of Dr. Paul Weigel at the University of Texas Medical Branch, Galveston, Dr. DeAngelis identified the first hyaluronan synthase to be described from the human bacterial pathogen Group A Streptococcus. Currently, as an Associate Professor at the University of Oklahoma Health Sciences Center in Oklahoma City, he directs a laboratory exploring various aspects of prokaryotic and eukaryotic polysaccharide biosynthesis. Discoveries from 1997 to 2000 include the identification of two more unique hyaluronan synthases and the first chondroitin synthase. Dr. DeAngelis co-founded a biotechnology company, Hyalose®, in 2000 to harness the basic science discoveries of polysaccharide synthesis. Raising orchids, thinking about natural and man-made antiquities, and scuba diving are his favorite pursuits when not playing with biomolecules.
Hyaluronan Synthases: Remarkable Catalysts
Hyaluronan is produced by many organisms and plays roles essential or very important for life. The enzyme catalysts that create the hyaluronan polysaccharide chain are called hyaluronan synthases [HASs]. HASs are efficient, dual-action glycosyltransferases that catalyze the reaction:
![]()
In contrast to the widely held belief that “one enzyme transfers one sugar,” the HASs co-polymerize both N-acetylglucosamine [GlcNAc] and glucuronic acid [GlcUA] into the hyaluronan chain. Elongation rates of 10 to 100 sugars per second have been measured in vitro. In all known organisms, the hyaluronan polymer is secreted or transported out of the cell upon synthesis, and the HAS activity has been found associated with the membrane fraction. The hyaluronan synthases from Gram-positive Group A and C Streptococcus bacteria and vertebrates have been studied for decadesa,b. In 1997-98, new hyaluronan synthases from an algal virus and a Gram-negative bacterium were identified and molecularly cloned.
a See review by Weigelin this series.
b See review by Spicer and McDonaldin this series.
Chlorella viruses, large double-stranded DNA (330 kb genome) viruses that infect certain Chlorella-like algae, are found in fresh water worldwide (Fig. 1)1. Virus titers can reach levels of 104 infectious particles per milliliter of water in ponds and rivers. These viruses were discovered initially by researchers pursuing the nature of symbiosis between algae and certain protozoa that comprise green paramecia or hydras. It was noted that the algal cells lysed hours after isolation from the paramecium or hydra. In 1981, Meints, Van Etten and colleagues used electron microscopy to reveal the presence of polyhedral particles in isolated algal cells, but not in the algal cells resident in the protozoa1. The laboratory host cells for Paramecium bursaria Chlorella virus (PBCV-1) are Chlorella-like symbiotic algae isolated from the protozoa. Within 3 to 4 hours after infection, the host lyses, and about 300 new virions are released. However, it is quite likely that the natural host is another, as yet unknown, free-living organism.
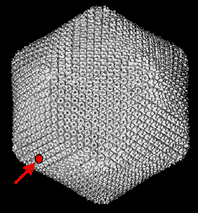
Fig. 1 Image of Paramecium bursaria Chlorella virus-1.
This computer reconstruction derived from electron micrographs depicts the virus that encodes a hyaluronan synthase, cvHas. The red dot is the size of a single subunit of the polyhedron coat. This large (175-190 nm diameter) Phycodnaviridae virus, which infects certain fresh water green algae, contains many types of genes that are not typical for a virus. Only the infected algal host cell, and not the virion itself, contains hyaluronan. (courtesy of Xiaodong Yan and Tim Baker.)
Similarity searches of DNA databases using the protein sequences of the streptococcal HAS (spHas or HasA) and the vertebrate HAS isozymes as queries suggested that an open reading frame, A98R (GenBank Accession # U42580), in the PBCV-1 genome was a potential HAS (Fig. 2). The viral protein contained seven short sequence motifs with similar placement within the polypeptide as the streptococcal and the vertebrate HASs. Interestingly, the PBCV-1 genome also encodes a functional UDP-glucose dehydrogenase that synthesizes UDP-GlcUA. Therefore, the virus does not need to rely on the host cell for this essential hyaluronan precursor. In 1997, the A98R open reading frame was amplified by polymerase chain reaction and cloned into an Escherichia coli expression system2. Biochemical analysis of the recombinant membranes for sugar transferase activity in vitro revealed that indeed a new HAS, now called cvHas, had been discovered. The cvHas is a very selective GlcNAc-transferase and GlcUA-transferase; other structurally related UDP-sugars do not substitute. Interestingly, manganese ion maximally stimulated HAS activity; in contrast, the streptococcal and the mammalian HASs prefer magnesium ion. The cvHas synthesized hyaluronan chains composed of ~104 sugars (3-6 x 106 Da) in vitro. Early after virus infection, the Chlorella contained cvHas messenger RNA and enzyme2. No enzyme was detected in the healthy Chlorella or the purified viral particles. Hyaluronan appeared on the infected Chlorella surface within 30 min post-infection, as judged by fluorescence microscopy utilizing an aggrecan detection reagent3. A further control, removal of the fluorescent signal by specific digestion by Streptomyces hyaluronan lyase, demonstrated that the infected cells formed bona fide hyaluronan. Electron microscopy of quick-frozen, deep-etched samples revealed that the infected Chlorella cells, but not healthy cells or hyaluronidase-treated infected cells, possessed an extracellular meshwork of intertwined fibers (Fig. 3)3. The discovery of cvHas was exciting for two reasons: (i) it is generally thought that viruses normally do not encode glycosyltransferases, but rather pirate the host enzymes to create viral glycoconjugates, and (ii) hyaluronan has never before been described outside of higher animals and certain bacterial pathogens.

Fig. 2 Sequence alignment of various Class I hyaluronan synthases.
The Chlorella virus enzyme (cvHas of PBCV-1) is very similar to the streptococcal (Group A S. pyogenes spHas) and vertebrate (human hHAS2) enzymes, as seen in this Multalin plot (90% consensus denoted in red; 50% consensus in green; Corpet F. Nucl. Acids Res. 16: 10881-10890, 1988). Seven short sequence motifs that are similarly spaced in the polypeptide are apparent. These regions probably play essential roles for the structure and/or function of these HASs because the motifs are present in all three types from disparate organisms.
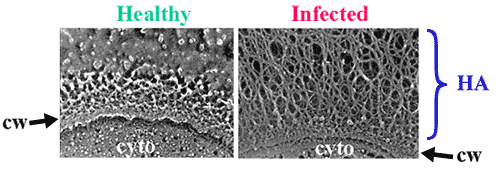
Fig. 3 Electron micrographs of healthy or infected algal cells.
These images reveal that the algal host cell develops an extracellular fibrillar coating after infection with PBCV-1. The fibers are composed of hyaluronan, as indicated by the interaction of infected algal cells with the hyaluronan-binding protein derived from aggrecan, or by fiber removal after treatment with Streptomyces hyaluronan lyase. The role of hyaluronan in the viral life-cycle is not clear, but since many isolates worldwide possess functional HASs, the role must be important. cw, cell wall; cyto, cytoplasm. (courtesy of Robyn Roth and John Heuser.)
Pasteurella multocida, a Gram-negative bacteria first described by Toussant and Pasteur in 1879-80, infects and often kills a wide range of animals, hence the origin of the species name4. Certain strains preferentially infect birds (fowl cholera), cattle (shipping fever), or swine (atrophic rhinitis), resulting in substantial economic losses. P. multocida is also found as an oral commensal organism in cats and dogs; consequently, this microbe is the major cause of sepsis in humans after inoculation via animal bites. The Carter group first recognized that certain fowl cholera isolates, classified as Type A, produced extracellular capsules that could be removed by treatment with hyaluronidase (Fig. 4). Chemical and NMR studies proved that the type A capsular polymer was indeed hyaluronan5. Biochemical analyses using native membranes prepared from a heavily encapsulated fowl cholera isolate showed HAS activity in vitro6. This enzyme activity also preferred manganese ion (like cvHas) instead of magnesium.
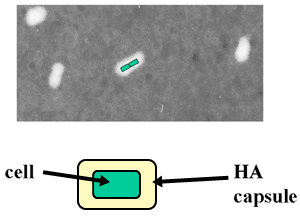
Fig. 4 Light micrograph of Pasteurella multocida Type A.
These Gram-negative rod-shaped bacteria often form pairs that are coated with hyaluronan. The hyaluronan capsule, which is readily visualized by negative staining with India ink, appears as a white halo around the microbes (cells depicted as green in one pair). The capsule serves as a virulence factor to enhance infection in fowl and bovine hosts.
Transposon insertional mutagenesis was utilized to tag and to identify the new HAS gene, named pmHas (GenBank Accession # AF036004), in 19987. Expression of this single 972-residue open reading frame conferred the appropriate E. coli strain with the ability to make hyaluronan capsules in vivo. Furthermore, membrane preparations containing recombinant E. coli-derived pmHas produced authentic hyaluronan polymer in vitro. Surprisingly, the pmHas amino acid sequence is quite distinct from the streptococcal, vertebrate, and viral HASs previously described. The pmHas is roughly twice as large and more closely related to other bacterial enzymes that synthesize distinct capsular polysaccharides and lipopolysaccharides (Fig. 5).
Several Pasteurella capsular types (B, D and F, in addition to A) with distinct polysaccharides have been described by Carter and other investigators at the U.S. Department of Agriculture (including Heddleston, Rimler, and Rhoades). The Type B polymer is composed of mannose, arabinose and galactose. Type D and F isolates produce heparin-like or chondroitin-like polymers, respectively. The first chondroitin synthase from any source was identified in a Type F P. multocida isolate by capitalizing on its similarity to the pmHas enzyme8. The new synthase, named pmCS, is 87% identical to pmHas; this close homology is understandable because the hyaluronan and the chondroitin backbones are structurally identical except for the substitution of N-acetylgalactosamine [GalNAc] for GlcNAc.
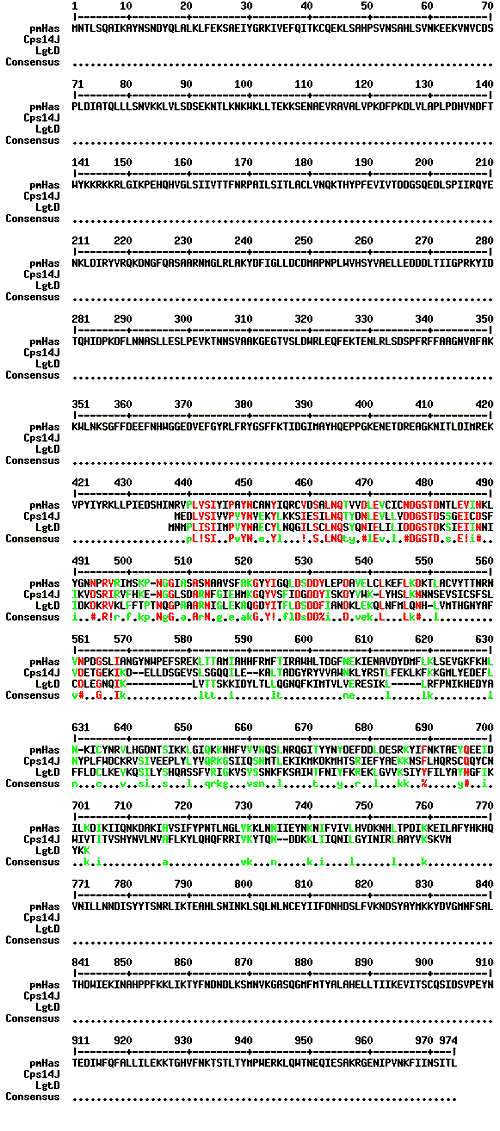
Fig. 5 Sequence comparison of Pasteurella pmHas and other glycosyltransferases.
This Multalin plot shows the similarity of a region of pmHas to other bacterial enzymes that produce distinct polysaccharides or lipopolysaccharides. All of these other enzymes (representatives shown here are Streptococcus pneumoniae Cps14J and Haemophilus influenzae LgtD) only catalyze the transfer of a single sugar linkage. The pmHas sequence does not align well with the other HASs (see Fig. 2) and, therefore, should be assigned to a separate class.
The role of hyaluronan in the Chlorella virus life-cycle is not known yet, but many isolates from around the world possess the HAS gene and cause their host to produce hyaluronan early in infection. Several hypotheses have been made, including: (i) prevention of secondary virus infection, (ii) stabilization of a fragile host cell to increase virus burst size, and (iii) facilitation of the interaction of the hyaluronan-coated infected cells with an alternative host organism2,3. At this point, some evidence indicates that the first scenario operates to a limited degree. A severe technical limitation is that the PBCV-1 system lacks the molecular genetic tools to create gene knockouts or specific mutants at this time. In light of the cvHas discovery and the amazing diversity of life, it is quite possible that other viruses in the aquatic or the terrestrial biosphere encode the production of hyaluronan or very similar polymers.
The extracellular bacterial capsule, in general, is thought to play one or more roles, including: (i) avoiding host defenses (e.g., complement and antibodies), (ii) facilitating adhesion or colonization, (iii) resisting desiccation, and (iv) modulating host physiology. Experimental evidence by various groups suggests that P. multocida uses the hyaluronan capsule for at least the first two roles. Due to the generation of capsule-specific antibodies, most capsular polysaccharides (other than hyaluronan) serve the pathogenic microbe only once. Hence, after a single infection, the animal is usually immune to infection by bacteria possessing the same capsular type. In contrast to many other anionic capsular polysaccharides, the hyaluronan polymer of Type A P. multocida is identical to the hyaluronan polymer of the vertebrate host. Therefore, this hyaluronan capsule is not immunogenic and, theoretically, is a better virulence factor because anti-capsule immunoglobulin defenses are not generated to repel a sustained or a second P. multocida infection. With respect to host colonization, it has been hypothesized that CD44, a prominent hyaluronan-binding protein in the vertebrate host, may serve as the anchor to localize hyaluronan-encapsulated Pasteurella and other Group A Streptococcus to certain tissues. Acapsular microbes that lack the hyaluronan capsule or bacteria stripped of their hyaluronan capsular coats by hyaluronidase treatment do not persist in the host or bind to isolated tissues.
Sequence and biochemical analyses of all known HASs suggest that at least two distinct HAS classes exist (Table 1)9. Enzymes from Streptococcus, vertebrates, and Chlorella virus (grouped as Class I HASs) possess seven short amino acid sequence motifs with conserved inter-motif spacing (Fig. 2). On the other hand, the Pasteurella HAS (the sole member of Class II) possesses variants of only two of these sequence motifs. Furthermore, Class I enzymes are predicted to be integral membrane proteins, whereas pmHas appears to interact indirectly with the membrane bilayer via an as yet unidentified partner (see Section IX). Therefore, it is likely that the two classes of HASs evolved independently to catalyze a reaction using the same UDP-sugar precursors to form the same polysaccharide. This hypothesis poses an interesting puzzle for biologists and enzymologists, as well as enhancing our respect for the importance of hyaluronan.
Table 1 Two Classes of Hyaluronan Synthases.
The Class I and II enzymes differ with respect to size, sequence, and predicted topology in the membrane. There may also be differences in the reaction mechanism based on the ability to elongate exogenously provided hyaluronan acceptors.
Class I
Members
spHas, seHas, cvHas
vertebrate HAS 1,2 & 3
pmHas
Polypeptide Size
417-588 residues
972 residues
Topology
multiple pass, integral membrane protein
soluble with a docking segment for membrane partner
Molecular Directionality of Polymer Growth
unclear (probably at reducing end))
nonreducing terminus extension
Acceptor Utilization
no acceptor found yet
yes; e.g., HA4
Based on the similarity at the amino acid level of pmHas to some lipopolysaccharide transferases, it seemed possible that hyaluronan oligosaccharides would serve as acceptors for GlcUA and GlcNAc transfer from their respective nucleotide sugars. Addition of unlabeled hyaluronan tetrasaccharide [HA4] derived from testicular hyaluronidase digests to reaction mixtures with recombinant E. coli-derived pmHas stimulated incorporation of radiolabel from UDP-[14C]GlcUA into polymeric hyaluronan by ~20- to 60-fold in comparison to reactions without the oligosaccharide10. On the other hand, structurally similar sugars (heparosan pentamer or chitotetraose) did not stimulate incorporation of the radiolabel. Gel filtration chromatography analysis of reactions containing recombinant pmHas, 3H-HA4, unlabeled UDP-GlcNAc and UDP-GlcUA showed that radiolabeled polymers from ~0.5 to 5 x 104 Da (25-250 monosaccharides) were made10. The activity of the native pmHas isolated from P. multocida membranes, however, was not stimulated by the addition of hyaluronan oligosaccharides under similar conditions. It is likely that the native pmHas enzyme has an attached or bound nascent hyaluronan chain that was initiated in the live bacterium prior to membrane isolation. The recombinant enzyme, on the other hand, is expected to lack a nascent hyaluronan chain because the E. coli host does not produce the UDP-GlcUA precursor. Therefore, the exogenous hyaluronan-derived oligosaccharide has access to the active site of recombinant pmHas and can be elongated (Fig. 6) 10.
On the other hand, neither recombinant spHas nor recombinant Xenopus xlHas1 (originally named DG42) that was prepared in Saccharomyces yeast (a host which cannot support hyaluronan biosynthesis) elongated hyaluronan-derived oligosaccharides into larger polymers10. This finding corroborates early reports by the Dorfman group utilizing native streptococcal enzyme. These recombinant preparations of streptococcal or vertebrate enzymes, however, were highly active in the conventional HAS assay, in which radiolabeled UDP-sugars were polymerized into hyaluronan. This difference is another line of evidence for the existence of two classes of HASs (see Section V).
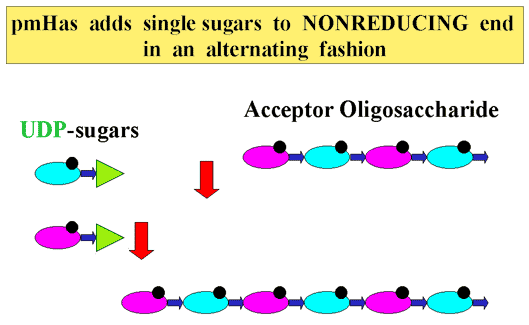
Fig. 6 Model of pmHas-catalyzed hyaluronan elongation.
Hyaluronan-derived oligosaccharides (HA4 depicted) can be elongated by recombinant pmHas in vitro. Single monosaccharides (GlcUA, magenta; GlcNAc, blue) donated by UDP-sugar precursors are added in a step-wise fashion to the non-reducing terminus of the acceptor hyaluronan chain. The alternating sugar-repeat structure of hyaluronan is generated by the sequential action of the two selective glycosyltransferase activities of pmHas.
The HA4 derived from ovine testicular hyaluronidase digests of hyaluronan terminates at the nonreducing end with a GlcUA residue and serves as an acceptor for hyaluronan elongation by pmHas. On the other hand, the delta-tetramer and delta-hexamer oligosaccharides produced by the action of Streptomyces hyaluronan lyase did not stimulate hyaluronan polymerization (Fig. 7). As a result of the lyase eliminative cleavage, the terminal unsaturated sugar is missing the C4 hydroxyl of GlcUA that would normally be extended by the hyaluronan synthase. A closed pyranose ring at the reducing terminal GlcNAc was not required by pmHas since reduction of HA4 with borohydride did not affect its ability to serve as an acceptor. Therefore, elongation of the nascent hyaluronan chain in Pasteurella occurs at the nonreducing terminus10.
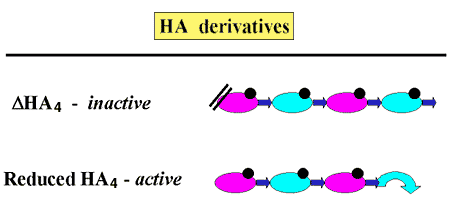
Fig. 7 Directionality of pmHas-catalyzed hyaluronan elongation.
The molecular directionality of hyaluronan elongation was confirmed by testing the ability of various HA4 derivatives to serve as acceptors. The presence of a dehydrated GlcUA at the nonreducing terminus (delta-HA4) prevented elongation, but opening the pyranose ring at the reducing end (reduced HA4) did not affect elongation.
If only the next appropriate UDP-sugar precursor was available in the reaction mixture, recombinant pmHas elongated the oligosaccharide substrate by a single sugar unit. For example, GlcNAc derived from UDP-GlcNAc was added onto the GlcUA residue at the nonreducing terminus of HA4 to form a pentamer. On the other hand, inclusion of only UDP-GlcUA did not alter HA4. If both hyaluronan precursors were supplied, then longer products were made. Thus pmHas transfers individual monosaccharides sequentially during the polymerization reaction. This reaction mode is similar to the action of the mammalian heparin and chondroitin synthases (or copolymerases). In Pasteurella hyaluronan biosynthesis, the repeating disaccharide structure of hyaluronan is generated by the fidelity of the two individual transferase activities (Fig. 6)10.
Much progress was made in 1999-2000 towards understanding hyaluronan biosynthesis due to the unique ability of the pmHas polypeptide to be assayed for the two half-reactions that comprise hyaluronan polymerization, either GlcNAc-transferase or GlcUA-transferase. The pmHas sequence has a duplicated ~100-residue element in the regions located at residues 161-267 and residues 443-547 (Fig. 8). These elements possess conserved sequence motifs containing DGS and DXD that are found in HASs of both Class I and II. We named these two elements of pmHas domain A1 and domain A2, respectively. This nomenclature is based on the similarity of these pmHas domains to the “A” domain proposed for other glycosyltransferases that make beta-linked carbohydrates11.

Fig. 8 Schematic model of the domain structure of pmHas.
Inspection of the deduced pmHas protein sequence reveals the presence of two repeated elements. Analysis of various truncation and site-directed mutants reveals that the first element, domain A1, contains a GlcNAc-transferase activity (blue) and the second element, domain A2, contains a GlcUA-transferase activity (magenta). Kinetic analyses suggest that the sites are relatively independent (e.g., a mutation in one site leaves the other site unaffected). The carboxyl terminal region contains a segment critical for interaction with the bacterial membrane (green).
The X-ray crystallography structure has been determined for another predicted glycosyltransferase, SpsA isolated from Bacillus subtilis, whose specificity is not yet known (!)12. The structure suggests that the aspartate residue in the DGS motif is involved in binding the UDP-sugar by interacting with the N3 group of the uracil ring, whereas the second aspartate residue in the DXD motif coordinates with the metal ion complexed to the phosphates of the UDP-sugar (Fig. 9). The conserved aspartates, residues 196 and 477, of the DGS motifs in the two putative domains of pmHas were mutated13. All of the pmHas mutants were either inactive or made long hyaluronan polymers with low efficiency as measured by the full HAS assay. However, pmHas domain A1 mutants (containing D196E, D196K or D196N substitutions) maintained high levels of GlcUA-transferase activity. On the other hand, pmHas domain A2 mutants (containing D477E, D477K or D477N substitutions) had high levels of GlcNAc-transferase activity. The pmHas aspartate mutants apparently lose their ability to bind their respective precursor sugars and, therefore, do not incorporate the monosaccharide into the hyaluronan chain. The mutant proteins with asparagine or glutamate substitutions at D196 or D477 were inactive or very poor transferases, indicating that both the size and the charge of the aspartate side chain of this motif are critical for interaction with uracil. This result greatly strengthened the hypothesis that two distinct transferase sites exist in the pmHas enzyme; domain A1 is essential for GlcNAc-transferase activity and domain A2 is essential for GlcUA-transferase activity (Fig. 8). The selective disruption of either the GlcNAc-transferase or the GlcUA-transferase does not significantly perturb the remaining transferase activity as assessed by the Michaelis constant values (KM). This finding suggests that the transferase domains are rather independent. Thus with further development, pmHas derivatives should prove useful as controllable biotechnology catalysts for the creation of specific oligosaccharides or hyaluronan-derived devices or molecules.
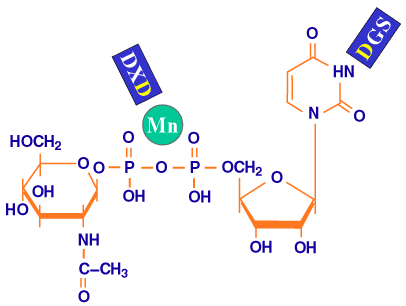
Fig. 9 Schematic model of the roles of the repeated motifs.
Based on the sequence similarity of pmHas and a protein of known three-dimensional structure, SpsA, the putative roles of the DGS and the DXD motifs (blue) are binding the uridine ring and coordinating the metal ion (green sphere) of the sugar precursor, respectively.
Further evidence for two independent active sites in the pmHas polypeptide was obtained from enzyme complementation in vitro13. The standard hyaluronan synthesis assay was performed with a mixture of the GlcNAc-transferase mutant enzyme and the GlcUA-transferase mutant enzyme. This mixture elongated hyaluronan-derived acceptors in a rapid fashion. The simplest explanation for this reconstitution of hyaluronan synthase activity in vitro is that the nascent hyaluronan chain is extended by one functional transferase, released, and extended by the other transferase in a repetitive, nonprocessive fashion. At this time, in contrast, the streptococcal and vertebrate enzymes, Class I HASs, are thought to polymerize a hyaluronan in a processive fashion with chain release only after chain completion a.
a See review by Weigel in this series.
With respect to the active sites of Class I enzymes, the Kimata group examined a solubilized murine Has1 enzyme that possessed both HAS and chito-oligosaccharide (short GlcNAc polymer) synthesis activities14. Mutation of aspartate residue 242 of the DSD motif resulted in ~99% reduction of both activities. Mutation of leucine 314 in the SGPLG motif disrupted the HAS activity, but the chito-oligosaccharide polymerizing activity was preserved. Therefore, it appears that two active sites also exist in Class I HASs, but the role of these particular motifs in hyaluronan polymerization is not yet clear.
Subcellular localization of various truncated pmHas mutants revealed that the carboxyl terminus is essential for targeting this HAS to the lipid membrane (Fig. 8)13. The mutated pmHas1-703, which lacks the last 269 residues, retained HAS activity but was transformed into a soluble cytoplasmic protein. The carboxyl-terminus, even though it does not contain any predicted hydrophobic or membrane-associated segments, appears to be responsible in large part for the association of native pmHas with the membrane by interaction with another bilayer-associated molecule. A potential partner is one of the proteins that compose the transport mechanism required to export the capsular polysaccharide in some Gram-negative bacteria such as Haemophilus and certain E. coli.
The Class I HAS enzymes, in contrast, possess 6 to 8 predicted membrane-spanning and membrane-associated regions in several regions of the polypeptidea,b. This model is in agreement with recent topological experiments performed on spHasa,15. It is quite likely that the Class I enzymes transport the growing hyaluronan chain through the membrane bilayer during biosynthesis, but testing this hypothesis is experimentally challenging.
a See review by Weigel in this series.
b See review by Spicer and McDonald in this series.
The two new HASs from Chlorella virus and from Pasteurella bacteria have certainly enhanced our understanding of hyaluronan biosynthesis. At least two classes of HASs exist: Class I, composed of the streptococcal, vertebrate, and viral enzymes, and Class II, composed only of pmHas thus far. The discovery of cvHas highlights the potential biological importance of hyaluronan in all sorts of life-forms, and it has provided a finer focus on the “critical” regions of HASs via sequence comparisons. In contrast, pmHas illustrates that nature can devise an alternative pathway to achieve the same goal of producing hyaluronan. The unique biochemical attributes of pmHas have revealed how the coordinated action of two exquisitely selective glycosyltransferases within a single pmHas polypeptide can build hyaluronan by alternate addition to the nonreducing terminus.
Many questions still remain. What are the amino acid residues and the structures responsible for the specificity of the transferase activities? What is the nature of the pmHas hyaluronan-acceptor site? What is its putative counterpart for retaining the nascent hyaluronan chain in Class I enzymes? How do the transferase sites of a dual-action enzyme encounter and elongate the growing hyaluronan chain? Elegance and grace will assuredly be components of the underlying mechanisms of the HASs.
Acknowledgments: I would like to thank Drs. Jim Van Etten and Michael Graves for providing images of the virus and infected cells. This work was supported by grants from the National Institutes of Health (R01-GM56497) and the National Science Foundation (MCB-9876193).