
Kiyoko F. Aoki-Kinoshita
Kiyoko F. Aoki-Kinoshita received her Ph.D. in computer engineering from Northwestern University in 1999. After a brief period as a post-doctoral fellow at the Institute of Information Science, Academia Sinica in Taiwan, she worked as a senior software engineer at BioDiscovery, Inc. in Los Angeles for three years. Then, from 2006, she moved to the Bioinformatics Center, Institute of Chemical Research, Kyoto University, where she started her research career in glycoinformatics. She is now a professor at Soka University, where she currently teaches and continues to do research to develop useful glycoinformatics tools for the community and to apply them to the understanding of glycan function in biological system.
This is the first of a series of articles on bioinformatics resources for glycoscience research. This first article will give a brief introduction to background knowledge that would be useful to read the rest of the articles. In particular, a description of the commonly used text and symbolic nomenclature to represent glycans will be given, along with recommendations for which representation to use. This is followed by brief descriptions of the types of resources that will be explained in this series. Glycan-related web resources vary from databases to web services to web portals of integrated data. It is difficult to try to weed through this web of data, and so this article attempts to organize the information in a way that users can understand the current state of these resources. Note that although there is a section on “glycoconjugates” which covers not only glycoproteins, but also glycolipids, proteoglycans, GPI-anchors, etc., the current state of databases only covers glycoproteins in general. The other types of glycoconjugates are not forgotten, however, and will be the focus of future development.
There have been many reviews in the literature about various glycan nomenclature, but here, we will provide recommendations as well.
There are four major glycan text formats that can be recommended for use by the community, due to their widespread use: CarbBank, IUPAC, GlycoCT and WURCS. CarbBank is the original format used in the database of the same name and is recommended by IUPAC. IUPAC, in turn, recommends three variations for human-readable representations of glycans. GlycoCT and WURCS are the main formats used by glycoinformaticians. Here, we propose that IUPAC condensed be used as a representation that is concise and human-readable. However, if accuracy is more important than human-readability, then GlycoCT or WURCS is recommended. Most recently, however, if the text representation is not necessarily required, then GlyTouCan (described later) identifiers (Tiemeyer et al. 2017b) can be used for specification of glycans without regards to format.
The original glycan text format that was devised for a glycan structure database was the CarbBank format, which displayed the 2-dimensional structure of glycans in typewriter font (same-width letters) text. Figure 1 illustrates an example of a glycan (https://glytoucan.org/Structures/Glycans/G02657AK) represented in CarbBank format. This format is human-readable but is difficult for a computer to parse because of the vertical bars, which cannot be quickly linked with the neighboring monosaccharide.
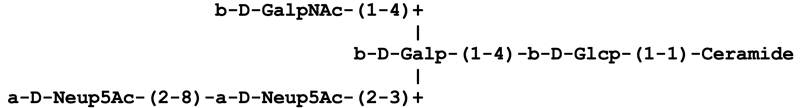
IUPAC is the International Union of Pure and Applied Chemistry, and they propose a nomenclature for representing complex carbohydrates called 2-Carb at https://www.qmul.ac.uk/sbcs/iupac/2carb/. In particular, section “2-Carb-38. Use of symbols for defining oligosaccharide structures” describes how to represent oligosaccharide structures by two basic recommendations: (1) The use of three-letter symbols for monosaccharide residues and (2) the 'reducing group', i.e. the residue with the free hemiacetal group or modification thereof (e.g. alditol, aldonic acid, glycoside), should be at the right-hand end, and when there is a glycosyl linkage to a non-carbohydrate moiety (e.g. protein, peptide or lipid) the glycosyl residue involved should appear at the right. Moreover, there are three recommended variations of carbohydrate nomenclature for oligosaccharides. These are summarized as follows:
Extended form: This is the format employed by CarbBank, where each symbol for a monosaccharide unit is preceded by the anomeric descriptor and the configuration symbol. The ring size is indicated by an italic f for furanose or p for pyranose, etc. The locants of the linkage are given in parentheses between the symbols; a double-headed arrow indicates a linkage between two anomeric positions. For example, α-D-Galp-(1→6)-α-L-Glcp-(1↔2)-β-D-Fruf represents raffinose.
Condensed form: This format is the most commonly used in various publications, databases and web pages as a rather compact but yet human-readable representation of carbohydrates. The configurational symbol and the letter denoting ring size are omitted, assuming that the configuration is D (with the exception of fucose and iduronic acid which are usually L) and that the rings are in pyranose form unless otherwise specified. Moreover, the anomeric descriptor is written in the parentheses with the locants. Reusing the example of raffinose, this structure would be written as Gal(α1-6)Glc(α1-2β)Fruf.
Short form: This form further omits locants of anomeric carbon atoms, the parentheses around the locants of the linkage, and hyphens. Branches can be indicated on the same line by using either parentheses or square brackets. Whenever necessary, configuration symbols and ring size designators etc. may be included, to make the notation more specific. Thus, in short form, raffinose could be represented as Galα-6Glcα-βFruf or Galα6GlcαβFruf.
GlycoCT is the format that was developed under the EuroCarb project, initially led by Dr. Claus Wilhelm von der Lieth of the German Cancer Research Center (Ranzinger et al. 2009). This format is currently most popularly used by bioinformaticians as it is the format used in GlycomeDB (Herget et al. 2008), one of the first integrative glycan structure databases, which has now been integrated into GlyTouCan (Tiemeyer et al. 2017a) , the international glycan structure repository, due to difficulties in maintenance. The structure in Figure 1 is represented in GlycoCT as shown in Figure 2.

On the other hand, during the initial development of GlyTouCan, discussions regarding which format to use for glycan representation raised several issues about GlycoCT. Because GlyTouCan was being developed using Semantic Web technologies, one of the first requirements for the glycan representation was that it be a linear string. Secondly, it needed to be able to represent ambiguous structures without the use of a library. For example, new substitutions on monosaccharides should be able to be represented regardless of whether the substitution was already known. Thus, modifications needed to be specified by their chemical (atomic) structure, instead of by name. At the time, no existing glycan representation satisfied these requirements, so the WURCS (Web3.0 Unique Representation of Carbohydrate Structures) was developed (Matsubara et al. 2017). WURCS specifies rules to ensure that all glycan representations are unique, thus allowing every glycan to be represented with a unique identifier. Figure 3 is an example of the WURCS representation; the residues in GlycoCT are shown in square brackets, followed by a slash “/” to list the linkages and their configurations. Note that the residues containing the text “NCC/3=0” represents the N-acetyled modification of the residue.
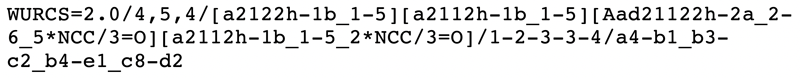
Because of the complexity of glycan structures, many glycobiologists use figures to illustrate them, and in addition to the CarbBank representation, currently there are two major ways to represent glycans using symbols: Symbol Nomenclature for Glycans (SNFG) and the Oxford notation. Although the SNFG has recently received much support from the community, it is a recommendation mainly for the symbols used to represent monosaccharides, whereas the Oxford notation also specifies the notation for glycosidic linkages to distinguish between anomeric forms and the carbon position. Thus, while the SNFG notation is recommended for monosaccharide symbols, the manner by which glycosidic linkages are represented is left to the user.
The SNFG symbol set was designed based on the discussions of a working group consisting of internationally recognized glycobiologists, chemists and informaticians. The list of currently recommended symbols is available at https://www.ncbi.nlm.nih.gov/glycans/snfg.html. It is based on the symbols originally recommended in the Essentials of Glycobiology textbook (Varki et al. 2017), which is often called the “CFG nomenclature”.
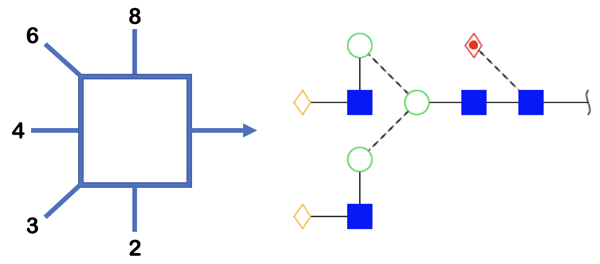
The Oxford system was designed by scientists at the University of Oxford Glycobiology Institute in 2009 using symbols that could clearly distinguish monosaccharides even using monochrome colors. Glycosidic linkages are represented as dashed lines for the alpha stereochemistry while solid lines are used for beta. Linkages are also drawn at angles to represent the carbon position where the bond is linked. Figure 4 illustrates the different angles used to represent the glycosidic bond conformation.
In this series, various glycan-related web resources will be described; in particular, those that can be accessed through the GlyCosmos Portal (https://glycosmos.org) will be introduced. In order to allow users to find their web resource of interest most effectively, we have categorized each database based on their biological contents, similarly to how GlyCosmos provides them. Similarly, here, each web resource will be briefly introduced, and the details of how to use each resource will be described in this series. Note here the difference between databases and repositories. Databases primarily consist of curated content that cannot be modified by users, whereas repositories allow users to add data in order to obtain a unique accession number. This distinction is being made in this series as well.
The genes that are most closely related to glycans are those that encode glyco-enzymes such as glycosyltransferases and glycosylhydrolases, involved in the synthesis of glycans, and genes that aid in the biosynthesis of glycans such as sugar-nucleotide transporters. The GlycoGene Database (GGDB) provided by ACGG-DB will be described in this series. Note that the Carbohydrate Active Enzymes (CAZy) database (Terrapon et al. 2017) in France is also very well known; collaborations between CAZy and GlyCosmos are ongoing in order to better integrate these data with other databases.
Glycan-related proteins can be categorized into the following based on how they interact with glycans: (a) protein products of glycogenes, (b) core proteins onto which glycans attach to form glycoproteins and proteoglycans, and (c) glycan-binding proteins which recognize and bind to glycans on the cell surface. Proteins in (a) would normally be found under Glycogenes; databases containing (b) include GlycoProtDB of ACGG-DB, UniCarbKB and GlyConnect, while UniProtKB and PDB also contain glycosylation data as well among their more comprehensive database of proteins; (c) includes the Lectin frontier Database (LfDB) of ACGG-DB, but data of glycan-array experiments can also be found in various databases developed in the US and Europe, which are currently being integrated by their respective projects. In the GlyCosmos Portal, these data would be found under the Glycoconjugates category.
Glycan-related diseases are either those caused by (a) mutations in glycan-related proteins encoded by glycogenes or (b) pathogens that interact with glycans and glycoproteins. Databases containing this information include the Glyco-Disease Gene Database (GDGDB) and PacDB, both provided by ACGG-DB.
Glycan databases were originally developed to provide additional annotations to glycan structures, such as pathways, diseases, etc. The international glycan repository GlyTouCan now is used as the central resource by which all glycan data can be linked, by using GlyTouCan IDs as accession numbers. GlyCosmos also provides access to the Total Glycome Database and GlycomeAtlas, both of which visualize glycomics data as obtained from mass spectrometry. Glycoconjugates refer to glycoproteins and glycolipids; glycoproteins are proteins which have glycans attached on them and glycolipids are lipids with glycans attached. Both are often found on the cellular surface. GlyComb is currently being developed as a repository for glycoconjugates, similar to GlyTouCan for glycans; it will serve as an important link between glycomics, proteomics and lipidomics. As a step towards this, GlycoPOST is now available as the mass spectrometry (MS) repository for raw MS data obtained from glycomics and glycoproteomics experiments.
In relation to glycan-related data repositories, there are currently two repositories that are available to the public, and one more under development at the time of this writing. GlyTouCan is the glycan structure repository, which provides unique accession numbers to any glycan entry, which can be a monosaccharide composition, a glycan with fragments, ambiguous linkages or monosaccharides, etc. Subsequently, anyone preparing to publish a manuscript about a glycan in any form can register it and obtain a unique accession number for it. The second repository currently available is GlycoPOST, which is a repository for mass spectrometry (MS) raw data for glycomics and glycoproteomics. In collaboration with UniCarb-DR and MIRAGE (see section later), GlycoPOST provides unique accession numbers to MS data and stores alongside it the metadata as recommended by MIRAGE.
As mentioned earlier, MIRAGE (minimum information required for a glycomics experiment) is an initiative to provide guidelines for reporting glycan-related experimental data. At the time of this writing, MIRAGE guidelines cover mass spectrometry (Kolarich et al. 2013), glycan arrays (Liu et al. 2017), sample preparation (Struwe et al. 2016) and liquid chromatography (in preparation).
In terms of informatics, while there are now numerous software and tools now available to analyze glycomics data, many are made publicly available in open source repositories such as GitHub. However, this makes it difficult for glycoinformaticians to find other resources. The Glycoinformatics Consortium (GLIC) was formed to provide a central repository of software, databases and developer information such that the community can find their software or database of interest. GLIC also provides a form for scientists to submit requests for software tools.。
GlyGen is a project funded by the U.S. National Institutes of Health Common Fund program that integrates various omics data with glycomics. Based on (glyco)proteins registered in UniProt, GlyGen displays glycoproteins, glycans and various proteoforms as published for human and mouse.
ExPASy is the Swiss Institute for Bioinformatics’ Bioinformatics Resource Portal which provides access to scientific databases and software tools (i.e., resources) in different areas of life sciences including proteomics, genomics, phylogeny, systems biology, population genetics, transcriptomics, etc. They have a page for Glycomics, which provides a list of internal and externally developed software and databases, to support glycomics research. One of their latest databases is GlyConnect, which aims to integrate glycomics and proteomics (Alocci et al. 2019) by visualizing glycoproteins, their site-specific glycosylation patterns, and the glycans that decorate them.
GlyCosmos is an integration project funded by the Japan Science and Technology Agency and the National Bioscience Database Center of Japan. Using Semantic Web technologies, glycans, their genes, proteins and related pathways and diseases are integrated and visualized in a user-friendly manner.
In the summer of 2018, these three projects (GlyGen, Glycomics@ExPASy and GlyCosmos) have agreed to share their data amongst each other, as the GlySpace Alliance. Because of the similarities and potential overlaps between these three projects, there was concern that users would be confused and split amongst them. In order to provide all data coherently, the GlySpace Alliance members have agreed to use open source licenses and software to share data between them. Annual meetings and discussions as to the various data that could be integrated will take place to ensure up-to-date and accurate data presentation.