
Kiyoko F. Aoki-Kinoshita
Kiyoko F. Aoki-Kinoshita received her Ph.D. in computer engineering from Northwestern University in 1999. After a brief period as a post-doctoral fellow at the Institute of Information Science, Academia Sinica in Taiwan, she worked as a senior software engineer at BioDiscovery, Inc. in Los Angeles for three years. Then, from 2006, she moved to the Bioinformatics Center, Institute of Chemical Research, Kyoto University, where she started her research career in glycoinformatics. She is now a professor at Soka University, where she currently teaches and continues to do research to develop useful glycoinformatics tools for the community and to apply them to the understanding of glycan function in biological system.
In this second installment of this series, an overview of the GlyCosmos Portal and the MIRAGE initiative will be described. The GlyCosmos Glycoscience Portal is a Web portal for glycoscience data resources, and as a member of the GlySpace Alliance, its data is openly available to the public. GlyCosmos provides access to glycan-related omics data, including glycogenes, glycoproteins, pathways, and diseases. Glycan-related repositories are also available, including GlyTouCan and GlycoPOST, for glycans and glycomics mass spectrometry experiments, respectively. MIRAGE is an initiative to provide standardized guidelines for reporting glycomics experiments. Because GlycoPOST closely works with MIRAGE, both of these are described in this installment.。
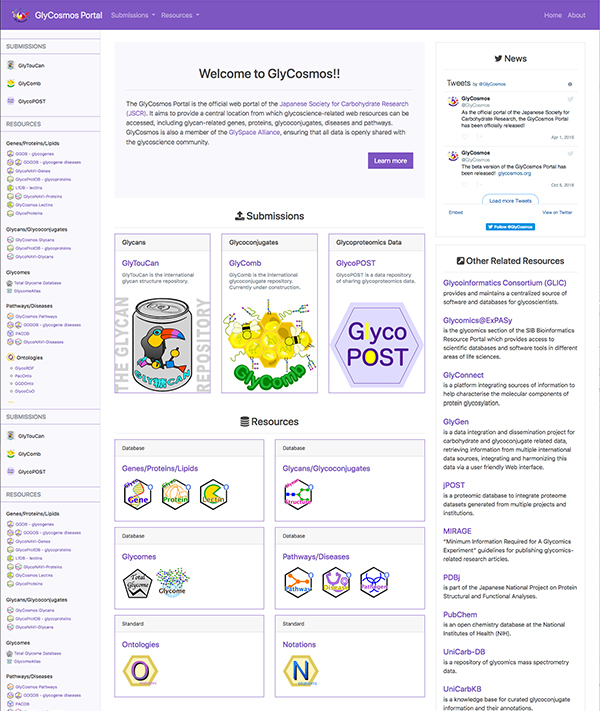
The GlyCosmos Portal is available at https://glycosmos.org. Released on April 1, 2019, it has been approved by the Japanese Society of Carbohydrate Research (JSCR) as its official portal for glycoscience data. In this section, the variety of resources available from this portal will be described.
The first repository ever developed for the glycosciences is the international glycan structure repository, GlyTouCan (Tiemeyer et al. 2017). Repositories differ from databases in that users can add information to the dataset in order to obtain an accession number for the data. Usually, repositories would allow the user to set a time for when the data would be made public, until which time the data would be stored privately and made available only to specific persons, such as journal editors and reviewers.
GlyCosmos provides access to three repositories: GlyTouCan, GlyComb and GlycoPOST. As mentioned above, GlyTouCan is the glycan structure repository. GlyComb is still under development at this time, but it is the glycoconjugate repository, and GlycoPOST is a mass spectrometry (MS) repository for glycans and glycoproteins. Each of these repositories will be briefly described in this section.
GlyTouCan provides accession numbers (formatted as the letter “G” followed by five digits and two alphabetical characters) to glycans, whether they are fully characterized with all known glycosidic linkages, or they are fragments, or they are just a set of monosaccharides (compositions). Users can register an account on GlyTouCan using their Google account, after which they have the possibility to register glycans using either the graphical tool, as text specifying glycan structures in either GlycoCT or WURCS formats, or as a file containing such text. Note that GlyTouCan accepts only structures with monosaccharides and their modifications; aglycons, or non-monosaccharide residues, such as amino acids must be removed before being registered. More details about how to use GlyTouCan will be described later in this series.
GlyComb is currently being developed as the glycoconjugate repository. No such repository currently exists, but there is a need to assign accession numbers to glycoconjugates and glycolipids. Therefore, GlyComb will provide such a system whereby users can specify glycans and the proteins or lipids they glycosylate. GlyComb will be available later this year, and so its details and usage will be described later in this series as well.
GlycoPOST was developed by the developers of jPOST (Okuda et al. 2017), a proteomics repository. It takes the information as specified by the MIRAGE guidelines (described later) for mass spectrometry data of glycomics experiments. Users create their own account using their email address, then create “Presets” containing metadata about their experiment as suggested by MIRAGE. Then they can link Presets with their Projects which contain the actual raw data. More details about the usage of GlycoPOST will be provided later in this series.
In GlyCosmos, each dataset listed below is displayed in their own subsection, annotated as either “Database” or “Standard” to differentiate between data and standards for representing the data. Icons (Table 1) indicate the type of data that can be found in each subsection.
Table 1: List of icons, the data they represent (Meaning) and selected resources in GlyCosmos that contain the relevant data.
| Icon | Meaning | Selected Resources |
 |
Glycogenes – any genes related to glycans, including glycosyltransferases, glycohydrolases, sugar transporters, etc. | GGDB, GDGDB |
 |
Glycoproteins – glycosylated proteins | GlycoProtDB, GlyCosmos Glycoproteins |
 |
Lectins – proteins that recognize and bind to glycans | LfDB, GlyCosmos Lectins |
 |
Glycans – carbohydrate sugar chains, usually with no aglycons attached | GlyTouCan (GlyCosmos Glycans) |
 |
Glycomes – the glycan-ome as characterized by mass spectrometry (MS) technologies of whole cells or tissues | GlycomeAtlas |
 |
Pathways – metabolic and signaling pathways in which glycoproteins or glycans are involved | GlyCosmos Pathways |
 |
Diseases – genetic and pathogenic diseases known to be caused by glycogenes or defects in glycan metabolism, etc. | GDGDB (glyco-disease gene database) |
 |
Pathogens – pathogens known to bind to glycans | PACDB |
Datasets (Resources) are organized by data type, and users can either click on a data type to go down the hierarchy or click directly on a specific data resource from the menu on the left-hand side of the page. Links to related resources are listed on the right. Each data type is described below.
At the time of this writing, the resources that are accessible from Genes/Proteins/Lipids are listed in Table 2. Currently, no lipid information is available, but glycan-related lipid data is currently being accumulated from the LIPID MAPS database (Sud et al. 2006). The majority of data resources in this subsection are provided by ACGG-DB (https://acgg.asia/db/) which is a portal for glycan-related databases in Asia. Each of these will be described in detail later in this series. The two resources provided by GlyCosmos are made possible by Semantic Web technologies (Aoki-Kinoshita et al. 2013), which allow us to integrate data from the Protein Data Bank (PDB) (Kinjo et al. 2018), UniProt (Bateman et al. 2017) and GlycoProtDB (Kaji et al. 2017). GlyCosmos Lectins is a list of protein entries in PDB that are annotated as lectins in UniProt. If a lectin is glycosylated, its glycosylation site information is also shown. For example, Polycystin-1 (UniProt ID P98161) is a heavily glycosylated lectin and can be found easily simply by sorting the list by number of glycosylation sites.
GlyCosmos Glycoproteins is a list of glycoproteins as annotated in UniProt. Combined with glycosylation site information from GlycoProtDB, each glycoprotein entry shows how each database has annotated their glycosylation sites. Since GlycoProtDB contains experimentally verified data, this information can be confirmed alongside the annotations from UniProt.
Table 2: A list of the data resources available under the Genes/Proteins/Lipids category
Each data provider is also listed and is accessible from GlyCosmos.
| Data type | Resource | Provider |
| Data type | GlycoGene Database (GGDB) | ACGG-DB | Glyco-Disease Genes Database (GDGDB) | ACGG-DB |
| Proteins | GlycoProtDB | ACGG-DB | Lectin frontier Database (LfDB) | ACGG-DB | GlyCosmos Lectins | GlyCosmos | GlyCosmos Glycoproteins | GlyCosmos |
Under the Glycans/Glycoconjugates section, users can access GlyTouCan and GlycoProtDB. As mentioned earlier, GlycoProtDB is a database provided by ACGG-DB and contains glycoprotein (glycosylated protein) information that has been verified experimentally using LC/MS-based technologies. Further information about the usage of these databases will be provided later in this series.
A glycome is defined as all of the glycans in a cell, tissue, or organism. Although high-throughput technologies to fully characterize glycomes is still at an early stage, many efforts have been made by groups around the world to do so. The GlycomeAtlas resource (Konishi and Aoki-Kinoshita 2012) was originally developed in RINGS (http://www.rings.t.soka.ac.jp) (Akune et al. 2010) and now provides a visualization tool for glycomes in human, mouse and zebrafish (Yamakawa et al. 2018). This resource is now also available from GlyCosmos.
The TotalGlycome database was developed to visualize the MS data as characterized by Furukawa et al. (Furukawa et al. 2017). It contains quantitative data from glycomics analysis of N-glycans, O-glycans, glycosphingolipids, glycosaminoglycans and free oligosaccharides. A variety of visualization tools are also available to allow users to compare the various data that has been accumulated.
This subsection of GlyCosmos provides access to GlyCosmos Pathways, Glyco-Disease Genes Database (GDGDB) and PacDB. GDGDB is also accessible from the Genes/Proteins/Lipids subsection as it is relevant in both. GlyCosmos Pathways is a collection of pathways in which glycoproteins are involved. The pathways have been accumulated from the Reactome database (Fabregat et al. 2018), and they are visualized using the Signaling Pathway Visualizer (SPV) tool (Calderone et al. 2018).
PACDB is the abbreviation for the Pathogen Adherence to Carbohydrate Database, provided by ACGG-DB. It provides information about diseases in the pathogenesis of which interactions between microbial glycan-binding proteins and glycans on the host play an important role. At the time of this writing, 446 microorganisms have been documented, and information about the glycans to which they do or do not bind are listed. Literature references are cited to the original publications in which the data has been reported. More information about how to use GlyCosmos Pathways and PACDB will be described later in this series.
Ontologies refer to the vocabulary that is used to describe data in a systematized manner. The most well-known ontology is the Gene Ontology (GO), which organizes genes according to their molecular function, cellular locations and biological processes. In the glycosciences, GlycoRDF was first defined as an ontology to describe glycan structures (Ranzinger et al. 2015). Although the name contains the term “RDF”, it is not actually Resource Description Framework (RDF), but rather an ontology that was developed to be able to describe glycans in RDF, or Semantic Web terms. GlycoRDF allows glycans, specified by GlyTouCan ID, to be annotated with publication information, the experiments used to characterize the glycan, whether it came from a biological source or was chemically synthesized, etc. The ontology used by GlyTouCan is based on GlycoRDF, and because many other glycan databases are also using GlycoRDF, Semantic Web queries can be made across these datasets in a straightforward manner. More information about Semantic Web technologies can be found in (Aoki-Kinoshita et al. 2013, 2015; Kawano 2017).
In GlyCosmos, the Ontologies subsection provides access to information about GlycoRDF, as well as PacOnto (the ontology developed to describe the data in PACDB) and GGDonto (the ontology developed to describe the data in GDGDB) (Solovieva et al. 2018). GlycoCoO (pronounced “glī-kō-koo”) is the glycoconjugate ontology, which has been developed to standardize the representation of glycoconjugates.
Notations were described in the first installment of this series, and this section of GlyCosmos provides access to the details about each notation that is recommended by GlyCosmos. This includes WURCS, GlycoCT and the Symbol Nomenclature for Glycans (SNFG).
MIRAGE stands for the Minimum Information Required for A Glycomics Experiment (York et al. 2014). It is a set of guidelines set forth by the MIRAGE commission in order to specify the minimum information that would be required when reporting a glycomics experiment, including mass spectrometry, glycan arrays, liquid chromatography, etc. MIRAGE is supported by the Beilstein Institut in Germany, and the MIRAGE commission is made up of well-known glycobiologists and glycoinformaticians from around the world.
One of the first MIRAGE guidelines that was proposed was for mass spectrometry experiments of glycans (Kolarich et al. 2013). This guideline provides a framework by which users can specify the necessary items to report when publishing their experimental results using MS for glycomics. UniCarb-DR (https://unicarb-dr.biomedicine.gu.se/) provides a Web tool by which users can enter their MIRAGE-related information for their experiment, and the user can thus obtain an Excel spreadsheet containing the information stored in a predefined format. In turn, GlycoPOST can import this information into a “Preset”, which stores the basic information about the MS equipment and parameters. Presets can then be stored and reused for all future MS experiments (assuming it does not drastically change), which would be linked with their “Projects” containing the raw data and peaklists.
Other MIRAGE guidelines are still being developed, and the interested user can access the MIRAGE home page at https://www.beilstein-institut.de/en/projects/mirage for the latest information.