
Kiyoko F. Aoki-Kinoshita
Received her Ph.D. in computer engineering from Northwestern University in 1999. After a brief period as a post-doctoral fellow at the Institute of Information Science, Academia Sinica in Taiwan, she worked as a senior software engineer at BioDiscovery, Inc. in Los Angeles for three years. From 2006, she moved to the Bioinformatics Center, Institute of Chemical Research, Kyoto University, where she started her research career in glycoinformatics. She is now a professor at Soka University, where she currently teaches and continues to do research to develop useful glycoinformatics tools for the community and to apply them to furthering the understanding of glycan function in biological systems.
In this final installment of this series, the possibilities of artificial intelligence applications to the various efforts around glycan-related database development are discussed. After a brief introduction to the field of AI, the compatibility of AI to glycobiology is described, followed by ideas for future research combining these fields.
Artificial Intelligence, or AI, is an all-encompassing term that covers a wide range of research topics, from robotics 1 to Web agents 2 to ubiquitous computing 2,3 to machine learning and deep learning 4. One of the areas of AI that is quite relevant to glycobiology is machine learning, and additionally deep learning.
While there are many machine learning methods depending on the type of learning (e.g., supervised and unsupervised) and data sets (e.g., classification, pattern prediction) at hand, in general, machine learning involves a procedure of processing large amounts of data under a particular model (e.g., text, graphs, images), in an attempt to “learn” high-probability patterns in the data. This learning process attempts to “train” on a set of data which is known to contain some commonalities, and it involves repeatedly adjusting the parameters of the model such that the target data can be predicted with high probability, or “accuracy”, oftentimes in comparison to a “control” data set. This is not necessarily easy, as many times the model has difficulty distinguishing between true positives and false positives. Moreover, there is the danger of over-training the model, called overfitting, which makes the model learn patterns that are too specific to the training data and cannot be generalized to other data sets. However, once a model is shown to be able to be generalizable with high accuracy, the model is considered to be “trained” such that it can make “predictions” on an unknown data set, such as whether it contains the pattern that it has been trained on, or whether it can be classified under a specific category. This is usually done by outputting a probability, or likelihood value.
One of the bottlenecks of such machine learning (and quite often artificial intelligence) models is that the trained model is usually a black box, in that the actual “meaning” of what has been trained is unknown. Recently, there has been work on making this box transparent (e.g., learning a white box, or interpretable machine learning) 5. As this book describes, interpretable models such as decision trees are of course possible, but these may not necessarily be very powerful or generalizable to large datasets of unknown data. Alternatively, a process called “feature extraction” is one way to make the black box of a trained model transparent, to produce the actual features of the data that has been learned. Such techniques will be important for the life sciences in order to be able to feed back to experimentalists the actual patterns that have been learned from the dataset. It would allow these experimentalists to more easily verify the prediction results. However, this is still an area of research that is ongoing in the field of machine learning, and in its infancy in the field of glycoinformatics.
Incidentally, glycobiology could also be considered to involve the process of trying to find meaning in ambiguity. This ambiguity lies in the ambiguity of glycan structures, where the structures are known to be rather flexible compared to other relevant biomolecules, such as proteins, and also include repeating structural units and a variety of modifications (e.g., sulfation, phosphorylation, etc.). There is also ambiguity in its recognition by lectins and enzymes, which could be mostly due to this structural ambiguity of glycans.
Because of this ambiguity in glycobiology, it seems to be a good target for applications of artificial intelligence and machine learning methods. In general, bioinformatics tools are often used to help screen experimental data to narrow down the options for the next steps in an experimental protocol. BLAST is an example of such a tool, allowing billions of sequences to be screened in a matter of minutes. As such, bioinformatics databases are useful for storing and organizing the large amounts of experimental data being produced by high-throughput experimental technologies. Subsequently, these databases are essential for machine learning and artificial intelligence techniques as they depend on large amounts of high-quality data. In terms of glycobiology data, it would be important to store glycan data describing their ambiguity and their recognition or binding affinity in order to be able to extract meaning from them. This can be done in terms of probabilities and statistics, as well as through standard nomenclature capable of encapsulating such information. Moreover, as the phrase goes, “Garbage in, garbage out,” it is also important to store high-quality and curated data in order to be able to effectively learn/train on the data to extract valuable information. Thus, standards are also important to ensure that the quality of the data is recorded along with the data itself (see the second installment of this series on MIRAGE). Indeed, the GlySpace Alliance has agreed to ensure that quality control is maintained across the alliance 6.
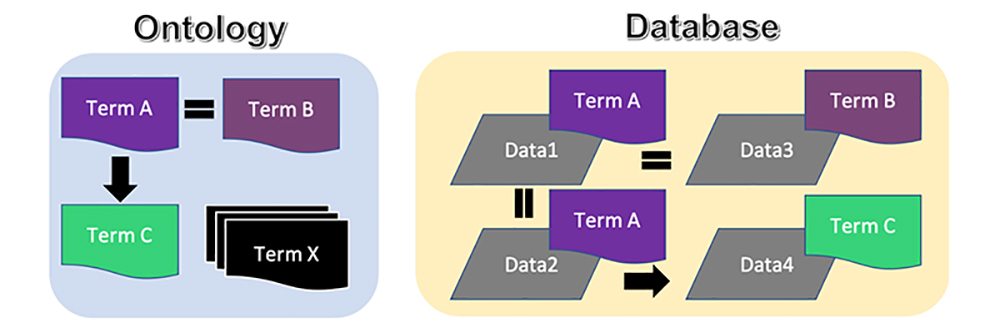
Semantic Web (SemWeb) technologies is one powerful way of storing standardized semantics with data, and its application to the life sciences has been described previously 7. Briefly, SemWeb involves the use of ontologies in order to define controlled vocabularies and their relationships in a formal manner. The Gene Ontology 8 is a well-known example whereby gene functions, their cellular localization and molecular properties are defined with identifiers for each and every term, organized in a hierarchical manner. Then, data are stored alongside these ontological terms such that the meaning of the data can be applied to them. Figure 1 illustrates ontologies, databases and their relationships. An ontology consists of terms to describe concepts that are encapsulated by data. These terms have relationships with one another, for example, in this figure, Term A and Term B are equivalent with perhaps minor differences in details, while Term C is a completely different concept that has some relationship with Term A (and Term B as well, as it is equivalent to Term A). There are many such terms and relationships defined as such in an ontology (Terms X in the figure). Now, on the right side of this figure, a database contains data that initially could not be interpretable by a computer (gray data); data are stored distinctly with no relationship between them. However, metadata can be attached to this data as ontological terms describing them. In the figure, Data1 and Data2 are thus defined as TermA concepts, while Data3 is TermB, and Data4 is TermC. With this additional information attached, the relationships defined in the ontology can consequently be inferred in the data. That is, both a human user and computer can deduce the relationships (black arrows and equal signs) between Data1 and Data2 (exactly the same concept), with Data3 (equivalent to Data1 and Data2), as well as Data4, being related to all three in some way.
By creating standardized ontologies that are used across databases, these semantics applied to data can be consequently standardized, allowing for easier integration of datasets. Thus the advantages of using SemWeb are that (1) disparate datasets across different servers and networks can be queried as an integrated dataset using the SPARQL query language, and (2) the computer can now be trained to infer new relationships in data across the Internet. In other words, what was previously called “islands” of data 9 can be integrated by implementing SemWeb technologies, and these data can be queried in a unified manner. Another recent advancement in SemWeb research is the development of similarity functions 10,11. The concept of “similarity” is normally difficult for a computer to handle, as it is built upon zeros and ones. However, it has been shown that correlations and similarity can be deduced between data using semantic analysis and ontologies 10. Thus, we can expect that such functions can be applied to glycan data, where concepts of ambiguity can be stored, processed, and even learned from the relevant data on the Semantic Web.
While there is a long way to go in terms of developing ontologies, constructing databases using these ontologies, and implementing computer models and methods to learn from these glycobiological data, it is possible to peek into the future paths available for glycoscience research. For example, it has been shown that it is possible to extract patterns of glycosylation across protein families. As protein glycosylation information have accumulated in major protein databases such as UniProt 12, machine learning methods have been developed to be able to make predictions on protein glycosylation sites 13–15. These can be further expanded with 3D data from the Protein Data Bank (PDB) to better understand the conformational patterns in substrates, or applied to other species other than eukaryotes, for example. In the near future, cross-species correlation analyses of glycan binding patterns, for example from glycan-array experiments, could be performed if these data are organized in a standard format across experimental technologies. In the past decade, glycan array technologies have advanced 16–21, producing large amounts of data about which glycans are recognized by particular glycan binding proteins, viruses, bacteria, etc. Many software tools are also available to analyze such data. However, there is no standardized and curated database of glycan binding patterns; this would be an important step for applying AI to understand glycan function. Furthermore, machine learning could be applied to the recognition process of 2D and 3D glycan structures by integrating such glycan array data with the 3D structures of the binding partners.
In the longer term, all these omics data, as integrated in GlyCosmos, can be utilized in Systems Glycobiology research, which could be categorized as follows: (1) Glycan biosynthesis simulations, (2) Cellular signaling by glycan binding simulations, and (3) Simulations of the extra-cellular matrix (ECM). For (1), AI could be used for predicting glycosyltransferase substrate specificity. One of the bottlenecks in glycan biosynthesis simulation is the definition of substrate specificities and the constraints on the substrates required for an enzymatic reaction to take place. A database of known specificities and the enzymatic properties of the glycogenes could be used to train a model to make such predictions. Similarly, the glycosyltransferase reaction parameters are also difficult to obtain, and thus machine learning could be performed on known data to make predictions on the values of Km, Vmax and other reaction parameters required for detailed simulation analyses.
In terms of cellular signaling and ECM simulations, these are research goals that currently have little to no systems glycobiology models currently available. It is well known in the glycobiology field that signaling events are oftentimes regulated by glycosylation 22–26 and that some glycosylation events in fact compete with phosphorylation to modulate signaling 27. Databases of these glycan-related signaling pathways are basically non-existent, let alone of signaling parameters. Thus, future work will entail the accumulation of such data and integration with the relevant omics datasets in order to be able to perform simulations and make predictions about signaling parameters. As for glycans and their functions in the ECM, it is starting to be better understood 28, but much still needs to be formally represented, such as the details about glycan structures (especially proteoglycans) in the ECM, their interactions with other proteins, and the configurations of these interactions. MatrixDB 29 is a starting point, additionally providing tools for network analysis. It can be expected to become important to further systems glycobiology research on the ECM.
Finally, as was described in the previous installment of this series on Cooperative organizations for glycoinformatics development, the Systems Glycobiology Consortium (SysGlyco) has been formed, with the aim to develop a database and tools for systems glycobiology research. SysGlyco currently consists of researchers developing such tools as GlycoSim and GlycoEnzDB. These are also starting points for furthering and integrating the relevant information needed to standardize and organize systems glycobiology data.
In summary, there is a lot of exciting research underway aiming towards concrete goals to better understand glycan function by applying AI and SemWeb technologies to glycoscience data. These are all efforts that require cooperation across researchers with diverse backgrounds and expertise; AI requires large quantities of data, which can only be produced by experimentalists. This is often not easy, due to language barriers between these disparate research fields. However, if such collaborations are successful, the results are potentially wide-ranging. Much focus is currently on medical fields, but other fields such as agriculture and energy are also interrelated with the glycosciences 30. It is my personal hope that such expansion of the glycoscience space can be made possible through these collaborations.