
Issaku Yamada
Received his Ph. D in engineering from Tokyo Metropolitan University in 1997. After having worked at Nagoya University (fellowship from Japan Society for Promotion of Science [COE program]), he became a Noguchi Institute researcher in 2002 and started scientific investigation of glycans at the Institute in 2006. He is presently engaging in glycan informatics studies on research topics such as glycan structure notation, ontology, and database design.

Kiyoko F. Aoki-Kinoshita
Received her Ph.D. in computer engineering from Northwestern University in 1999. After a brief period as a post-doctoral fellow at the Institute of Information Science, Academia Sinica in Taiwan, she worked as a senior software engineer at BioDiscovery, Inc. in Los Angeles for three years. From 2006, she moved to the Bioinformatics Center, Institute of Chemical Research, Kyoto University, where she started her research career in glycoinformatics. She is now a professor at Soka University, where she currently teaches and continues to do research to develop useful glycoinformatics tools for the community and to apply them to furthering the understanding of glycan function in biological systems.
In this eighth installment of this series, we introduce cooperative and organizational efforts to approach challenges in the glycosciences that would be difficult to solve by individual researchers, labs, or projects.
A number of countries have carried out glycan-related research and development projects thus far, and the glycoinformatics components of these projects have developed a variety of databases. However, the termination of these projects has led to the closing of websites and blocked utilization of the developed databases, which is an obstacle to glycan research. To solve this problem, glycoinformatics researchers all over the world together designed a system to share databases and software tools which cooperatively, has led to the establishment of the Glycoinformatics Consortium (GLIC). This consortium was initially supported the Noguchi Institute.
To contribute to the advancement of glycoscience by providing and managing a repository of glycoinformatics-related software and databases, and by creating a platform for glycoinformaticians, glycochemists, and glycobiologists to exchange information.
GLIC is creating a researcher-friendly environment by producing catalogs of glycan-related databases and other tools, and by providing and managing this repository. In addition, GLIC holds use-case workshops and hands-on seminars. These activities are open to the public at the website (https://glic.glycoinfo.org).
Working groups to develop glycan symbol nomenclature, ontologies and semantic web technologies have been formed. Online meetings were convened for the purpose of promoting the GLIC software repository, validating the reliability of the software and databases, creating nomenclature documentation, and evaluating validation methods. The documents and notes are disclosed on the website. Furthermore, GLIC holds use-case workshops on glycoscience research and collects and analyzes the results to contribute to the advancement of glycoscience.
Since the development of CarbBank 1, a number of glycan-related databases have been developed, and each database stores a unique meta-dataset on glycan structure, localization, and function. Development of the international glycan structure repository, GlyTouCan 2 greatly contributed to the ease of sharing of glycan information among researchers and databases. In August 2018, an alliance of three major glycan-related projects (Glycomics@ExPASy in Switzerland 3, GlyGen in the United States 4, and GlyCosmos in Japan 5) was formed, called the GlySpace Alliance to avoid duplication of research activities and project development 6.
The foremost goal of the GlySpace Alliance is to provide the scientific community with reliable information about biologically relevant glycans, including structure, glycan-containing glycoconjugates, biosynthesis, and roles of glycans in biological processes.
In order to participate in the Alliance, members need to have secure funds for development, maintenance and promotion of their projects. In addition, each member is also required to furnish valuable data, provide data use feedback (access logs, etc.) to evaluate the needs of users, and attend Alliance meetings.
Meetings are held once a year, gathering members in one place, but can also be online as needed. Topics generally discussed at meetings are data sharing and standard information. Data used in member projects are required to meet data standards; for example, UniProtKB 7 or RefSeq 8 are used for protein sequence identification, and GlyTouCan 2, PubChem 9, or ChEBI 10 are used for glycan structure identification.
The GlySpace Alliance is also planning to hold training sessions, in which members teach users how to use their portals and to guide users on how to carry out analyses on their individual data in a hands-on manner. Details are on the GlySpace Alliance website.
The GlySpace Alliance was established as a worldwide alliance to share glycan-related datab, which is an achievement comparable to the International Nucleotide Sequence Database Collaboration (INSDC), UniProt (a consortium of amino acid sequence databases), and wwPDB (a database of protein 3D structures). It is anticipated that the establishment of this Alliance will lead to unification of glycan-related databases all over the world.

The goal of the Systems Glycobiology Consortium (https://sysglyco.org/) is to develop an open platform for research into systems glycobiology, which requires information on signaling pathways and genetic interactions as well as glycosylation (biosynthesis of glycans).
Specifically, the Consortium aims to gather systems glycobiology and related researchers in order to be able to provide a collective resource including the following:
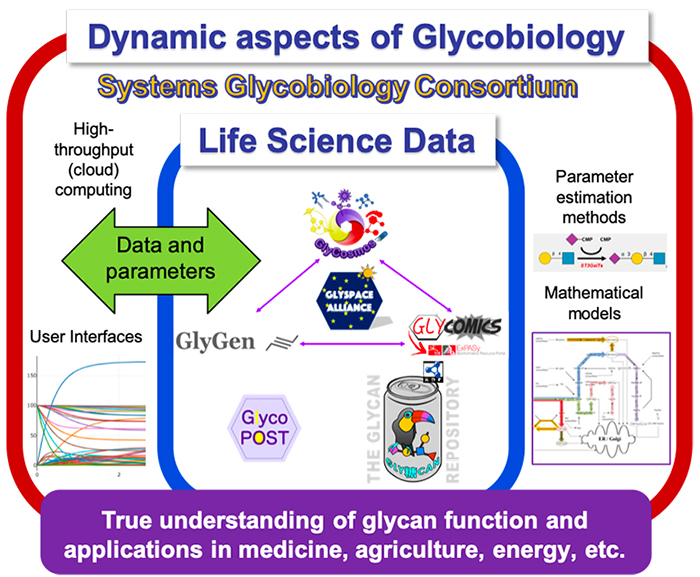
This Consortium was established at the end of 2019 and is now in its preliminary phase, but a mailing list has been created to provide an academic exchange platform for researchers worldwide in the field of systems glycobiology. Members of the Consortium will compile a database of reaction parameters and models required to build mathematical models, and once such data of reactions and glycan substrates can be standardized, it is anticipated that comprehensive analysis of glycan function could become possible.