Issaku Yamada
Dr. Yamada completed his doctoral studies at the Tokyo Metropolitan University Graduate School of Engineering in 1997. After working at Nagoya University as a Japan Society for the Promotion of Science special fellow (COE) and elsewhere, he was appointed as a researcher at the Noguchi Institute in 2002 and began to study glycoscience there from 2006. Since then, he has been engaged in glycan informatics research, including glycan structure notation, ontology development, and databases.

Kiyohiko Angata
After completing his doctoral studies at the University of Tsukuba Graduate School of Life and Environmental Sciences, Dr. Angata studied glycobiology at the La Jolla Cancer Research Foundation (presently the Sanford Burnham Prebys Medical Discovery Institute) under the tutelage of Professor Minoru Fukuda. In his current position at the National Institute of Advanced Industrial Science and Technology of Japan, he is conducting research to analyze glycan gene expression and new glycan functions, to find commercial applications of glycans, and to develop a glycan-related database (ACGG-DB).

Yu Watanabe
Yu Watanabe was appointed as a technical assistant at the Niigata University Graduate School of Medical and Dental Science in 2013 and has since served as a researcher. She has been in her current position as a specially appointed junior lecturer since 2018. She is now working to develop web-based databases and analytical tools for the life sciences.

Tamiko Ono
Tamiko Ono joined the JST Integration Promotion Program as a research assistant for the Kinoshita Lab in the Soka University Faculty of Science and Engineering in September 2017 and has been engaged in developing the GlyCosmos portal and collecting relevant data.
In this sixth installment of the series, we would like to describe databases and repositories for glycolipids and glycoproteins, which are collectively known as glycoconjugates. Many proteins are glycosylated and correspondingly exhibit a wide variety of functions, and these protein-modifying glycans comprise a wide variety of structures. It is also known that the proteins are modified with different glycan structures depending on diseases and other factors. Here is an introduction to databases for glycoconjugates and mass spectrometry data repositories for glycans and glycoproteins.
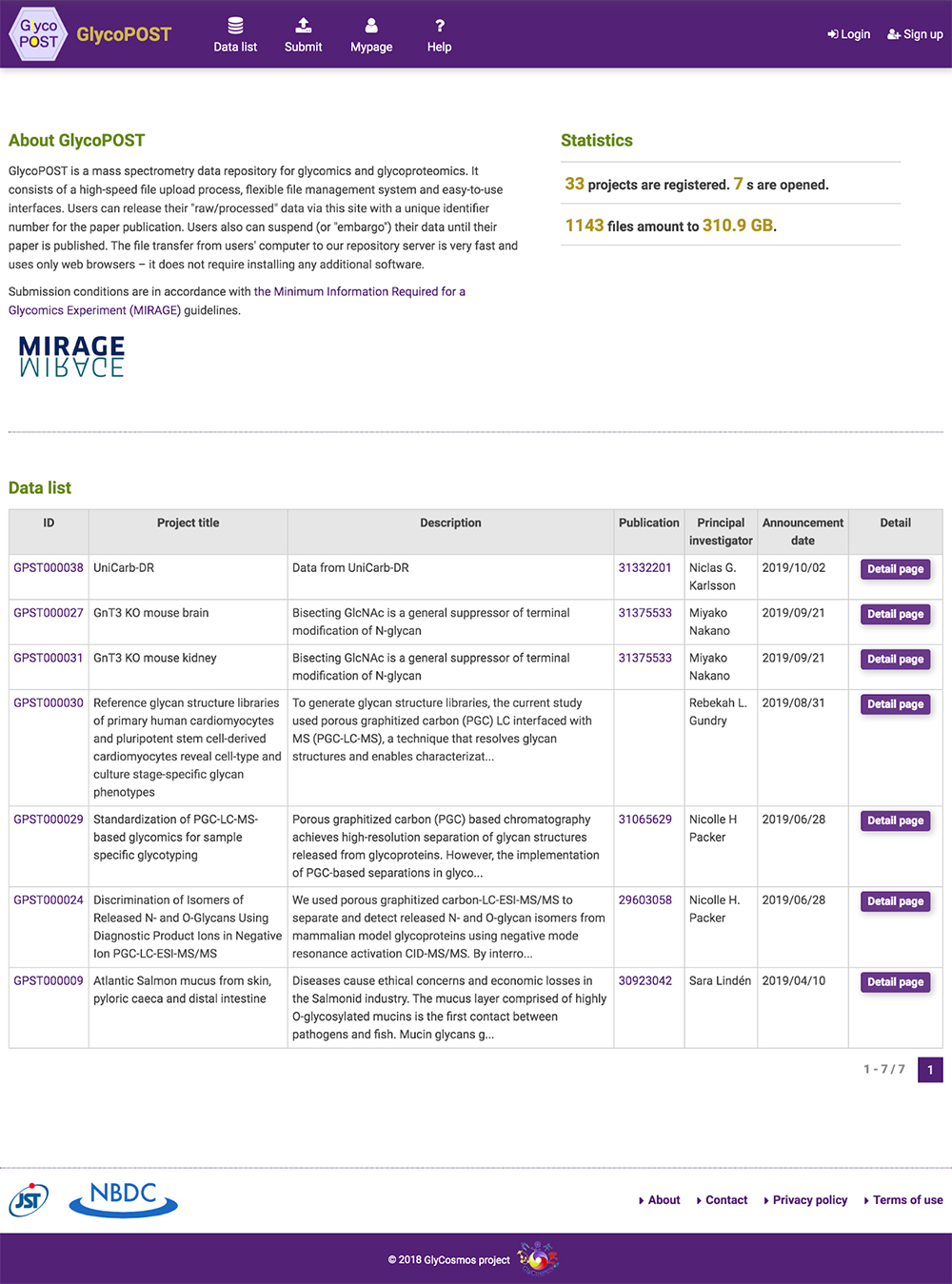
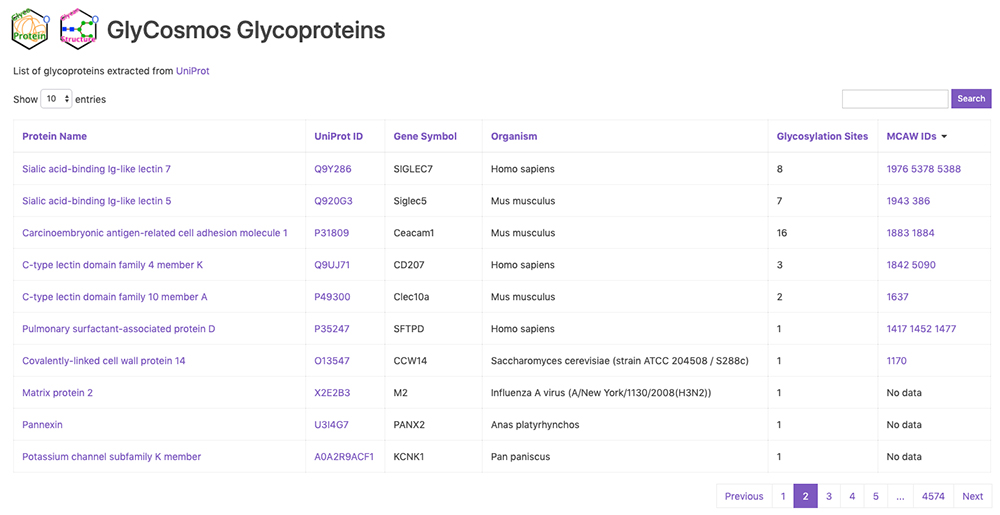
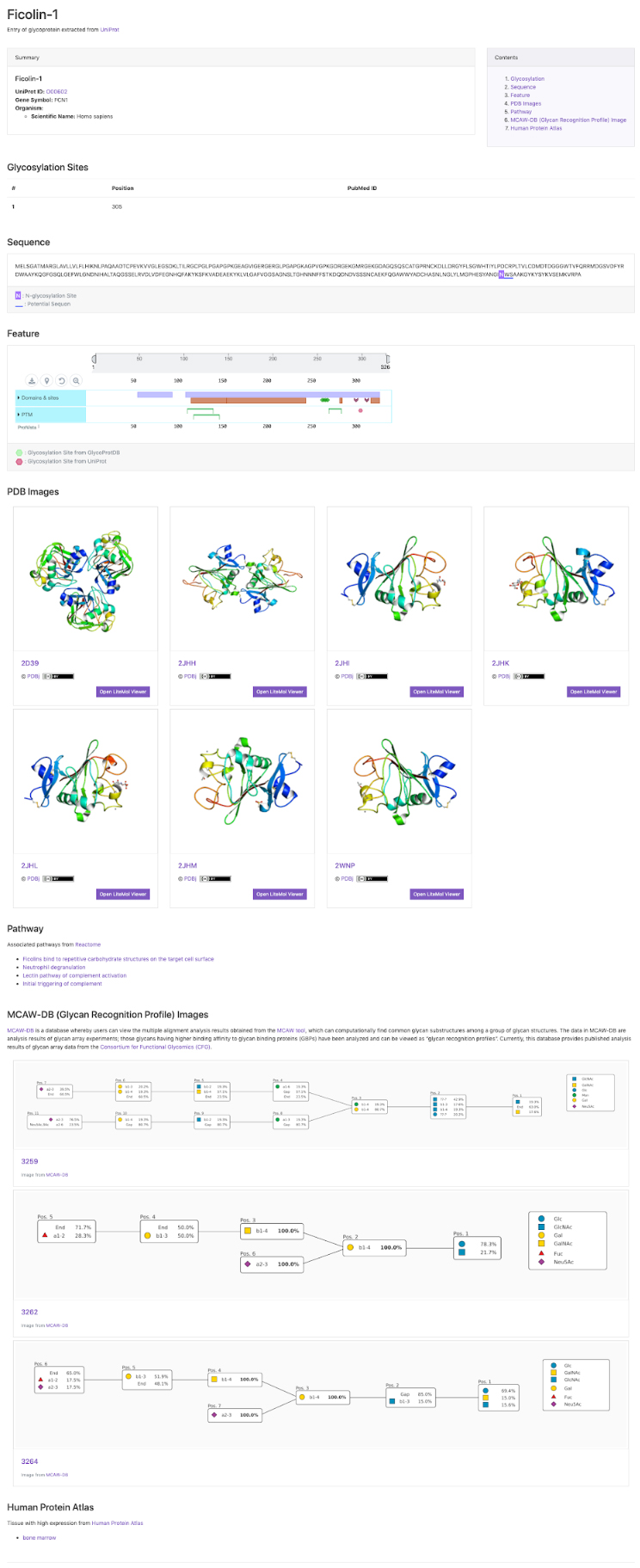
GlyCosmos Glycoproteins is a list of glycoproteins selected as proteins with glycan modification annotations from UniProt 1 (Fig. 1). Shown in the list are protein names, UniProt ID, gene symbols, organisms, and the number of glycosylation sites. MCAW IDs as linked to UniProt IDs in the MCAW-DB 2 are also displayed. Information on the MCAW-DB is available in the “Present Status of Lectin Databases” installment of this series. Protein Name and Gene Symbol text search is available by clicking on Search on the upper right side of the list. The Glycoprotein entry page (Fig. 2) can be accessed by clicking on the Protein Name in the list. The entry page has a wide variety of contents, including Glycosylation Sites, Sequence, Feature, PDB Images, Pathway, MCAW-DB (Glycan Recognition Profile) Image, and Human Protein Atlas (Table 1).
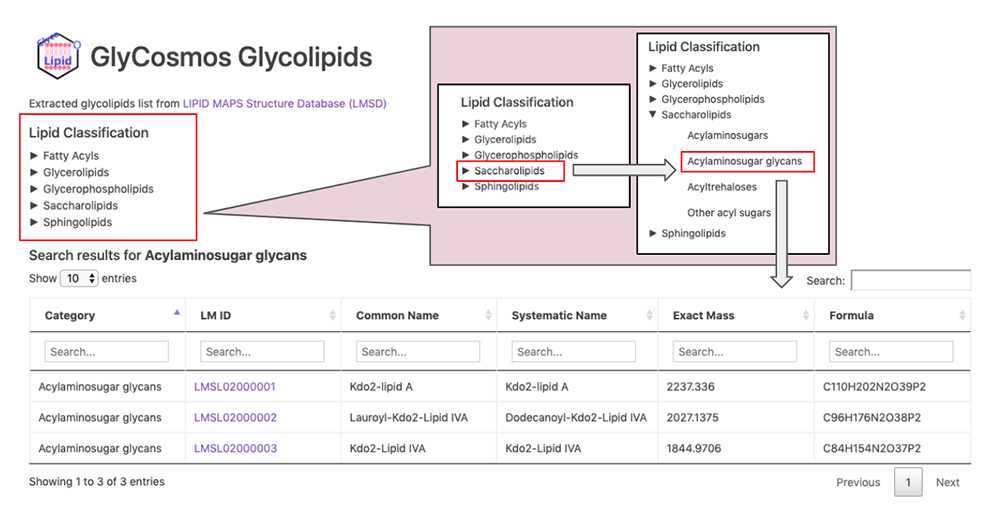
GlyCosmos Glycolipids is a list of glycolipids selected from the LIPID MAPS Structure Database (LMSD) 8 by keyword search for “glyco,” “glycan,” “sugar,” and “saccharolipids.” Select the desired category of glycolipids via the Lipid Classification on the GlyCosmos Glycolipid top page (Fig. 3). The categories are stratified; when the lowermost category is clicked, a list of glycolipids appears. Shown in the list are the category, LIPID MAPS ID (LM ID), common name, systematic name, exact mass, and chemical formula of each glycolipid. Clicking on an LM ID opens the corresponding LIPID MAPS page.
| Content | Description |
| Glycosylation Sites | Glycan modification sites, and, if available, PubMed IDs for literature information, are displayed. |
| Sequence | N-glycosylation Site and Potential Sequon information, along with sequences, are displayed. |
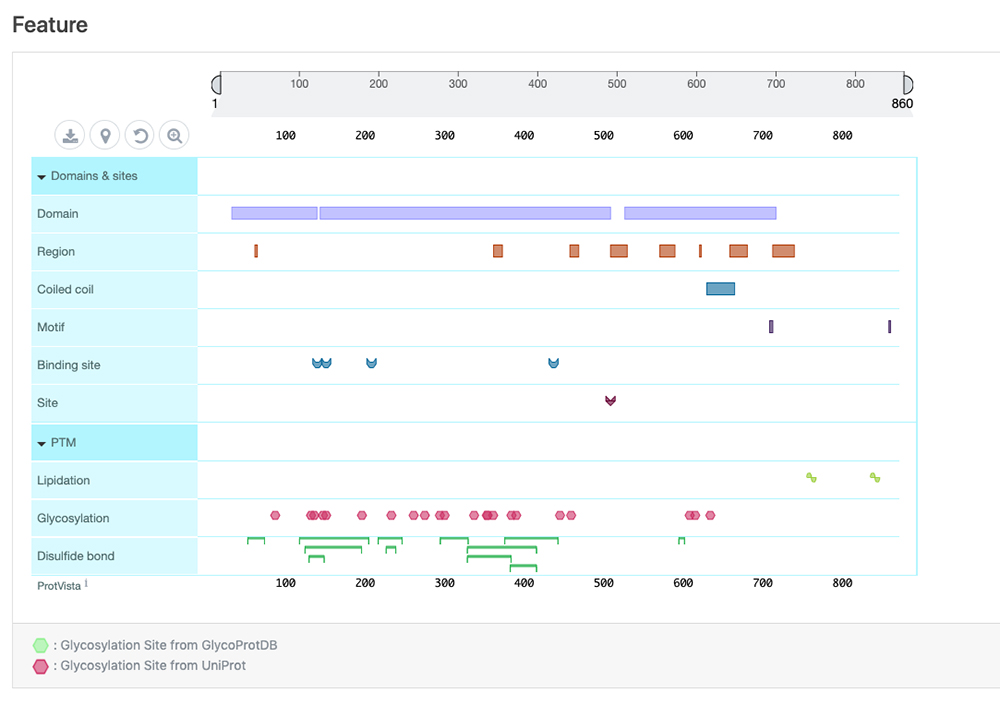
| Feature | Information on sequence annotations, including protein domains and amino acid modifications can be visualized. This display is powered by the ProtVista tool 3 (Fig. 4). |
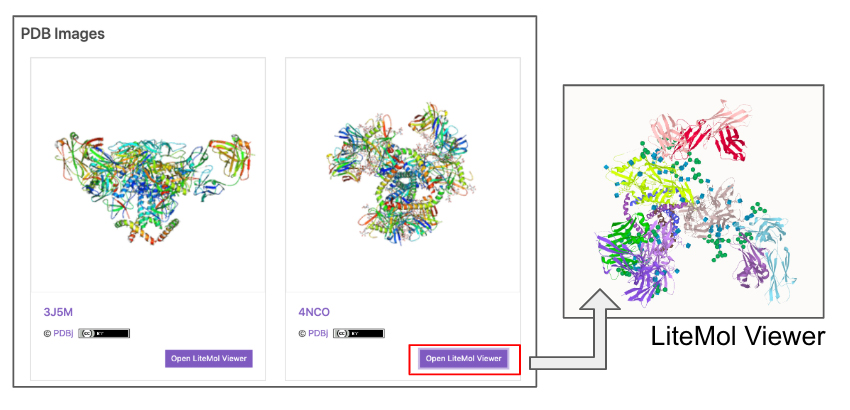
| PDB Images | Images of conformations from the Protein Data Bank (PDB) 4. A link to the LiteMol Viewer for 3D visualization 5 is available to allow molecular structures to be identified in more detail (Fig. 5). The LiteMol Viewer shows glycan structures in SNFG format. |
| Pathway | A list of pathways for which reactions are mediated by glycoproteins is displayed. The pathways shown here have been extracted from the Reactome database 6. When the desired pathway is clicked, the GlyCosmos Pathways page is opened and detailed information on the pathway can be found. |
| MCAW-DB (Glycan Recognition Profile) Image | MCAW-DB alignment results are displayed. |
| Human Protein Atlas | When the target species is Homo sapiens, organ-specific cell localizations can be realized. This screen shows a list of organs with “High” expression levels in the HumanProteinAtlas 7, and a link to the HumanProteinAtlas is available. |
In recent years, many studies have reported on various protein databases, including modifications, with the advances in proteome analysis and compilation of large data sets. However, only a few databases include glycan binding sites or glycan structures. Information is available from databases, such as Unipep for peptides with identified N-glycans; UniProtKB, which includes glycan modification site data; GlyConnect, which includes glycan compositions; and other databases. The Glycoprotein Database (GlycoProtDB, Kaji et al. 2012) has been open to the public within the framework of the Japan Consortium for Glycobiology and Glycotechnology Database (JCGGDB), providing N-linked glycosylation sites identified by mass spectrometry with a focus on glycan structures. At present, the latest version with modified interfaces (modes of display, etc.) is available in ACGG, the features of which are described below.
The GlycoProtDB lists data acquired from tissues, cells, sera and other biomaterials prepared from nematode, human, and mouse. The desired tissues and cells can be selected from the column on the left side, and displayed in alphabetical order (Fig. 6). In addition, data search can be achieved by entering the name of the glycoprotein of interest.
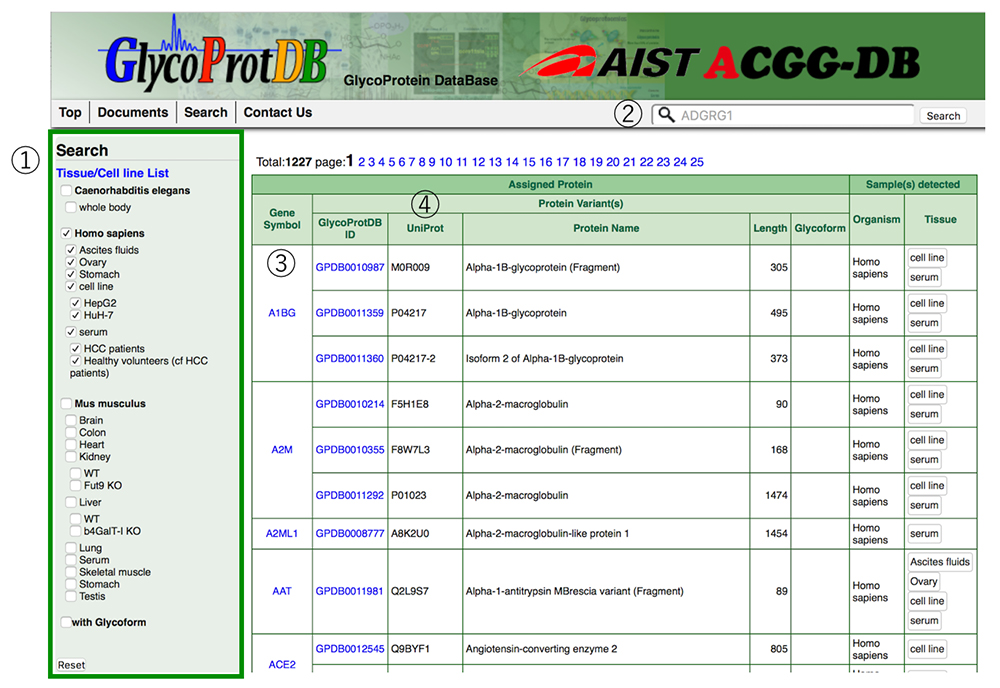
When a glycoprotein is selected from the search results, the N-glycosylation sites and amino acid sequence on the glycoprotein can be viewed on the detail page (Figs. 7 and 8). A major feature of GlycoProtDB is capable to find tissue- and lectin-binding-specific differences.
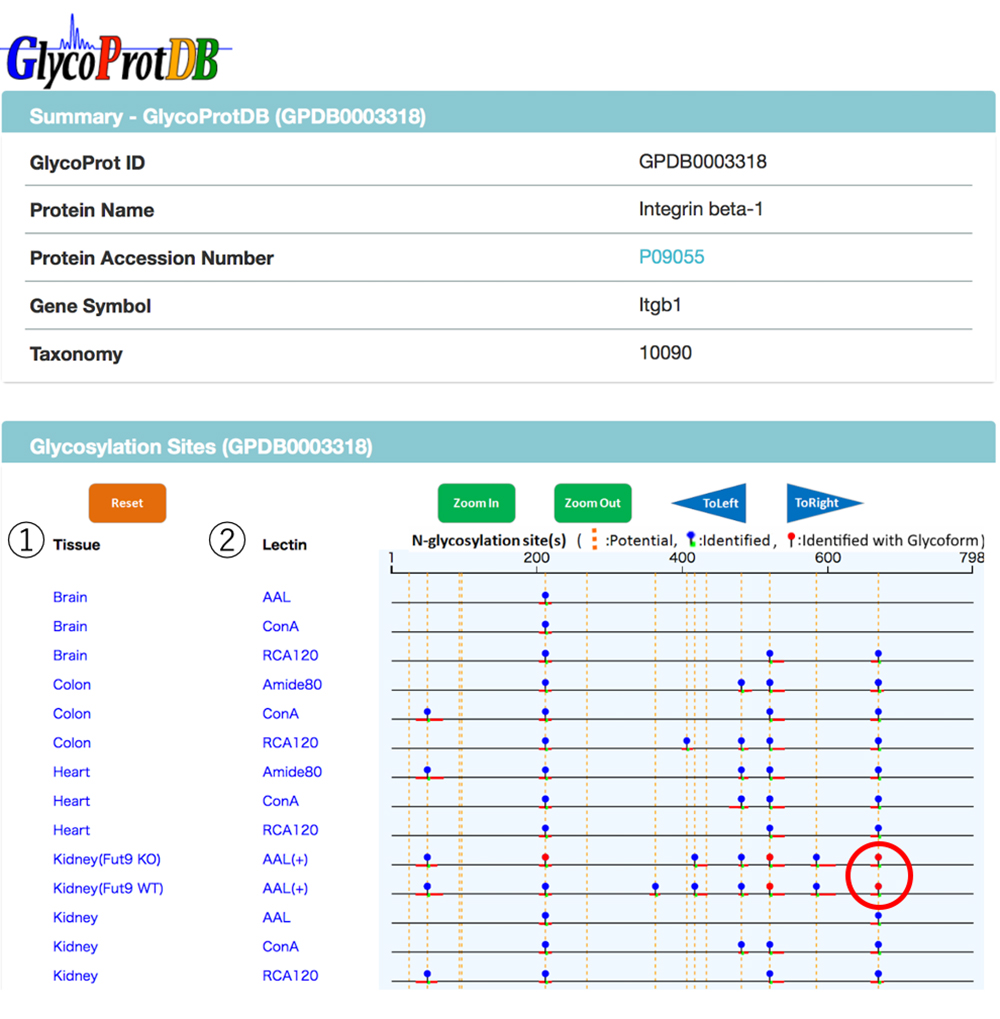
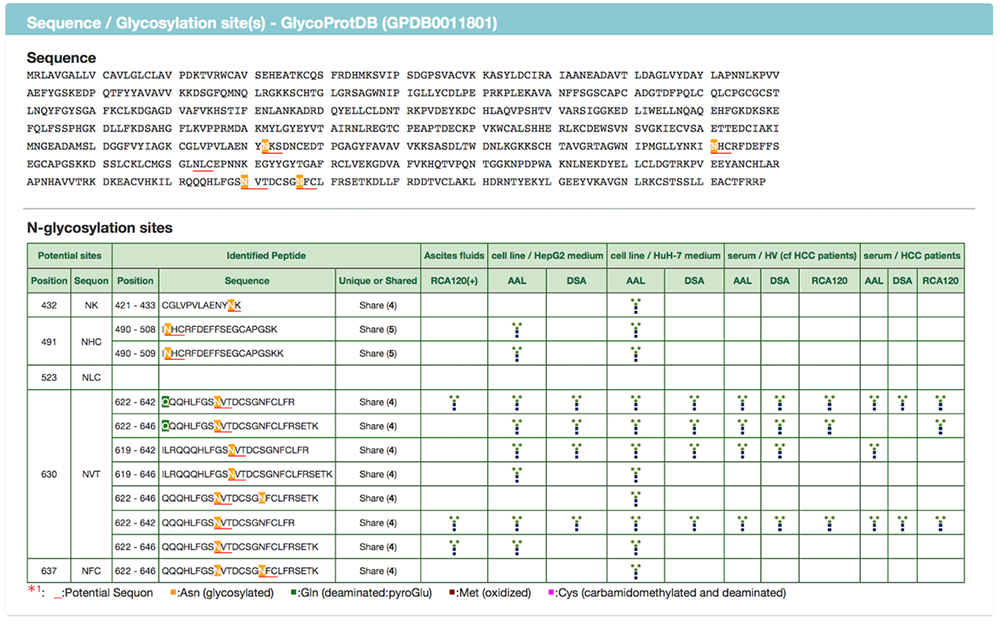
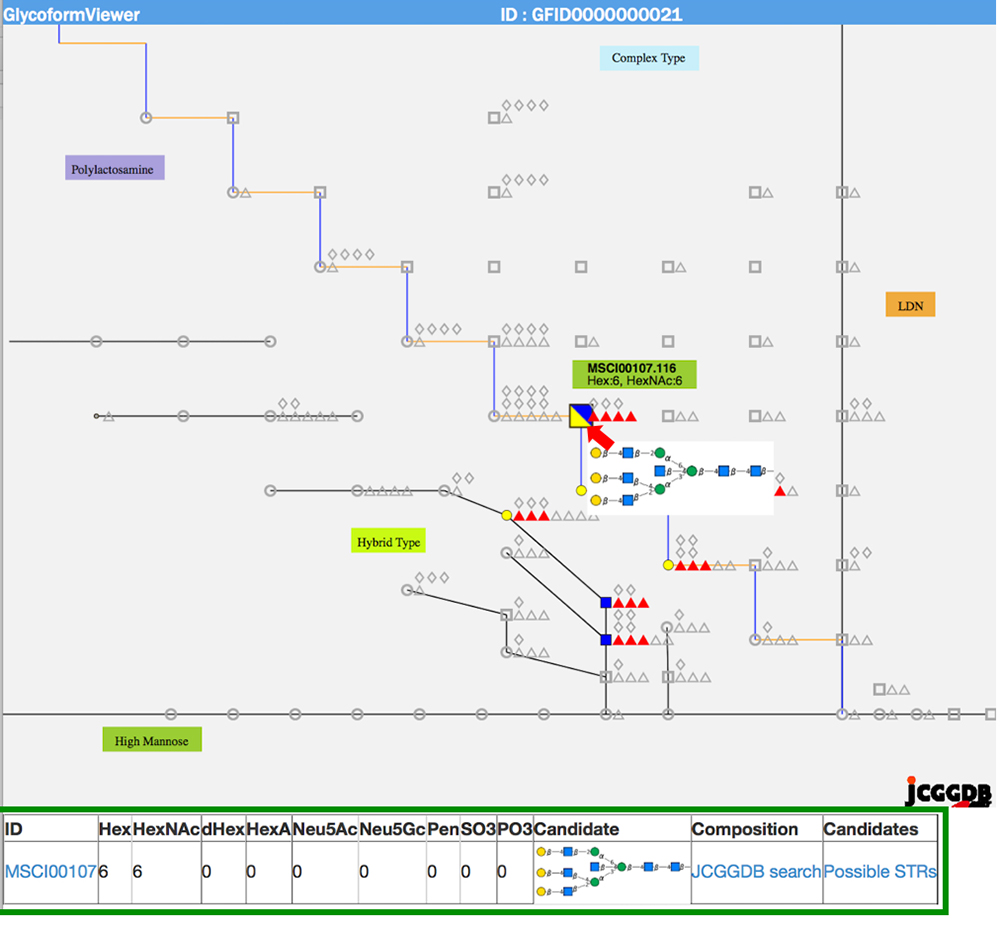
When multiple sites having glycan structures recognized by the same lectin are compared among different samples, tissue-specific differences can be seen. The results of an analysis of glycans obtained without cleaving from the glycoprotein (GlycoRidge method) are displayed with red pins (Fig. 7). When a red pin is selected, the glycan structure (glycan composition) can be found on the viewer (Fig. 9). When the cursor is moved onto the glycan identified in the viewer, candidate glycan structures are presented. Hence, the GlycoProtDB database not only makes it possible to know the true N-glycosylation sites, but also allows users to easily visualize tissue-, cell-, and serum-specific differences, including glycan structures.
GlycoNAVI is a website constructed to support glycan science research. On this website, data in the Protein Data Bank (PDB) 11 is analyzed, and a secondary database of organized glycan-related data (TCarp) is described. The PDB has data on conformations of glycoproteins, glycolipids, free glycans, and other entities. The list of glycan structures shown in Figure 10 can be accessed via the Glycans site on the GlycoNAVI top page. Glycan structures can be searched by sequentially accessing the list or entering the glycan structure repository GlyTouCan 12 accession number or a WURCS glycan structure representation 13. When a GlyTouCan accession number is clicked, the GlyTouCan entry page appears.
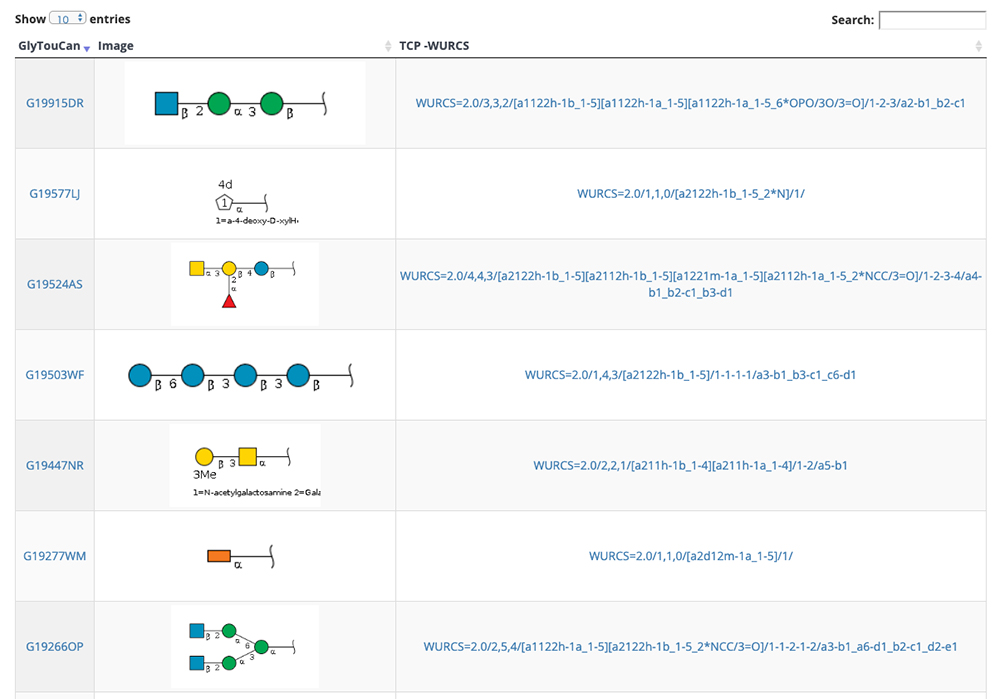
When the WURCS strings for the desired glycan structure in Figure 10 is clicked, a list of entry pages including the glycan structure (Fig. 11) appears. In this list, conformations, GlyTouCan accession numbers, and glycan structure SNFG representations (https://www.ncbi.nlm.nih.gov/glycans/snfg.html) 14 are displayed. Just as described above, the number of entries on one page can be changed via the upper left pulldown menu and search can be initiated by clicking on the Search button in the upper right. When an ID in this list is clicked, detail pages are displayed.

The list of glycan structures in Figure 12 can be accessed via the Proteins site on the GlycoNAVI top page. This list displays the number of glycans contained, PDB title, and other information. Just as described above, you can change the number of entries on one page via the upper left pulldown menu and search can be initiated by clicking on the Search button in the upper right. When an ID in this list is clicked, detail pages are displayed.

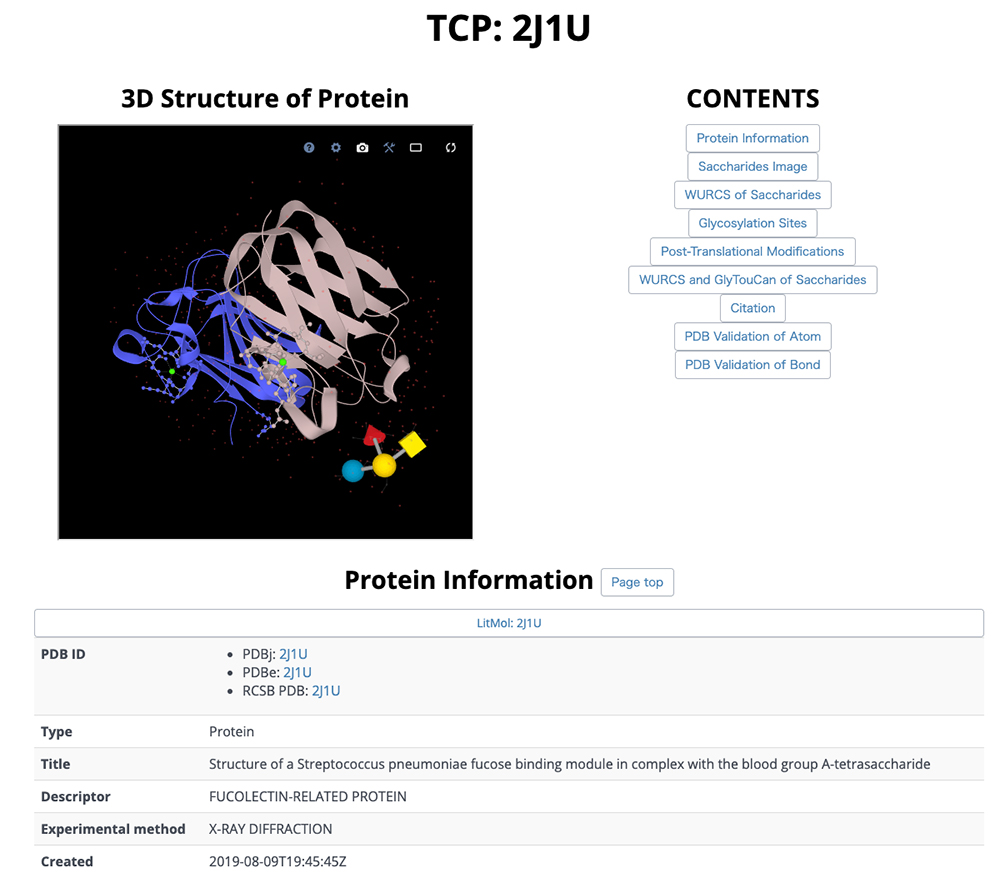
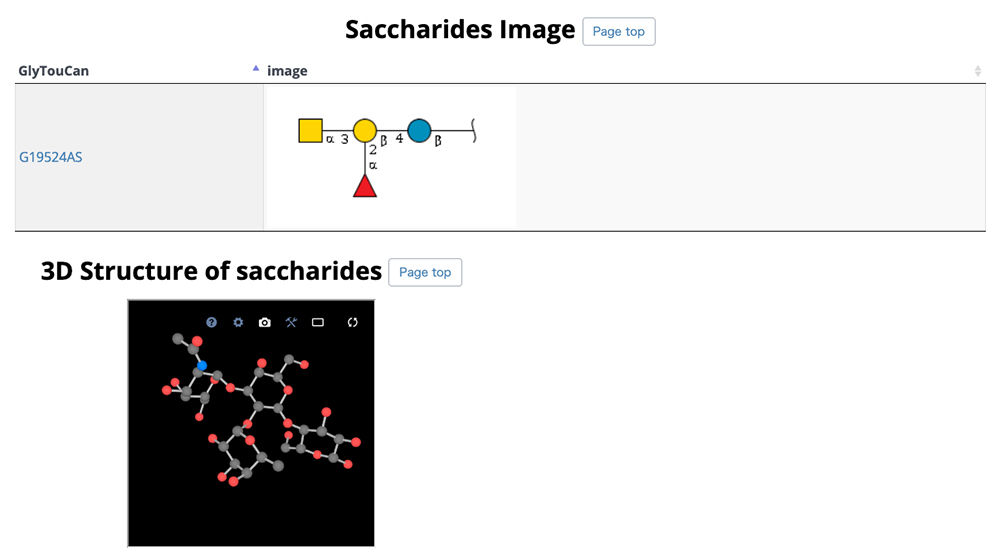
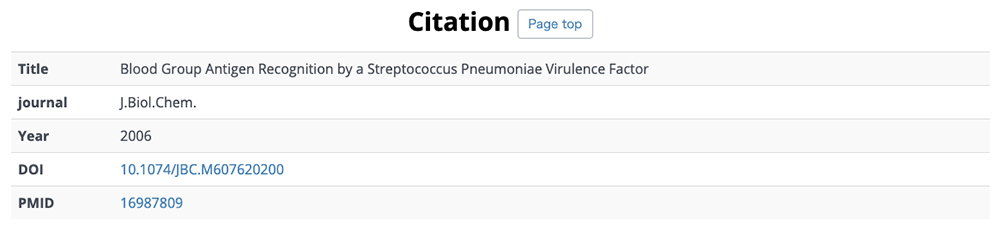
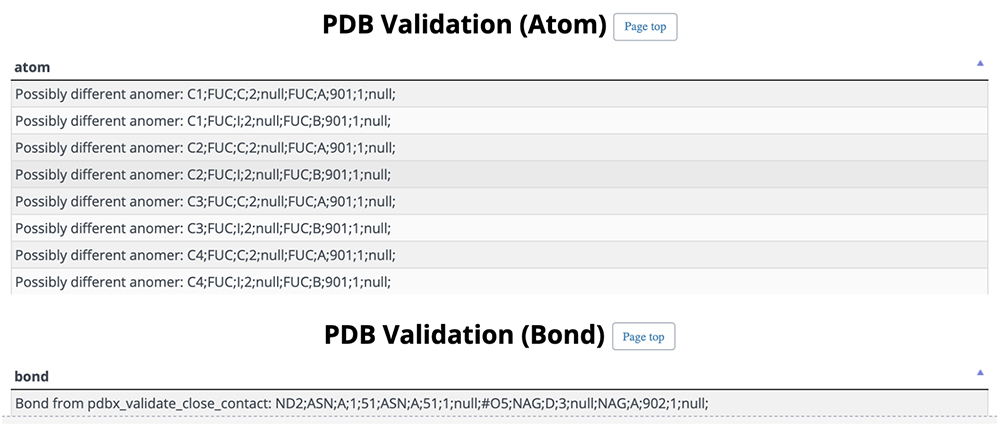
On this detail page, three-dimensional structural representations of glycan molecular structures are depicted using 3D-SNFG 15, PDB links, PDB entry titles and explanations, experimental procedures, analysis dates and times (Fig. 13), GlyTouCan accession numbers and links, glycan structure SNFG representations, glycan conformations (Fig. 14), literature references, PubMed links, digital object identifiers (DOIs), and their links (Fig. 15). Figure 16 shows the results of a glycan-related verification of chemical structure data analyzed. When displayed, this result means that there is any site to note in structures of the glycan.
GlycoPOST is a repository for depositing glycoprotein mass spectrometry data (Fig. 17). The user can register his/her own experimental data and access and download data registered by other users. This is often used to present experimental data relevant to a published article.
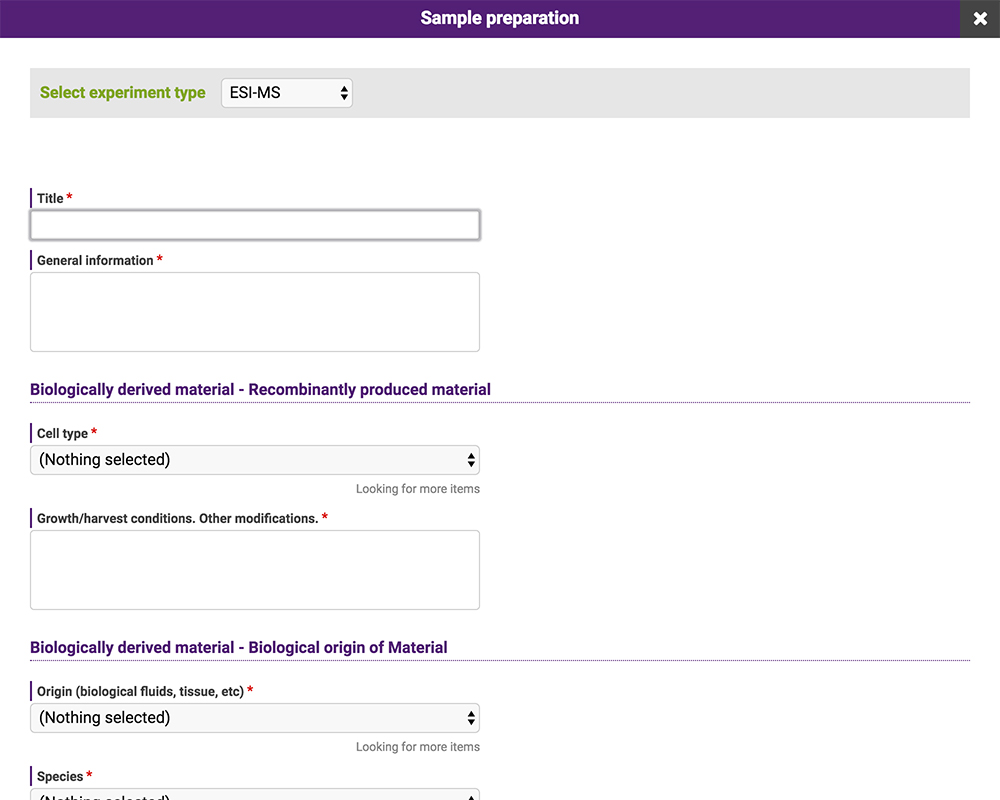
The data posted to the repository consist of meta-data, including experimental conditions, and electronic files. Meta-data can be entered in GlycoPOST by selecting the appropriate entry in the pulldown menu or providing a statement in the text box (Fig. 18). These entries comply with the guidelines for reporting glycan-related experiments proposed by MIRAGE 16. This database is compatible with other databases and repository sites that are in compliance with the same guidelines; data can be imported and exported via Microsoft Excel files.
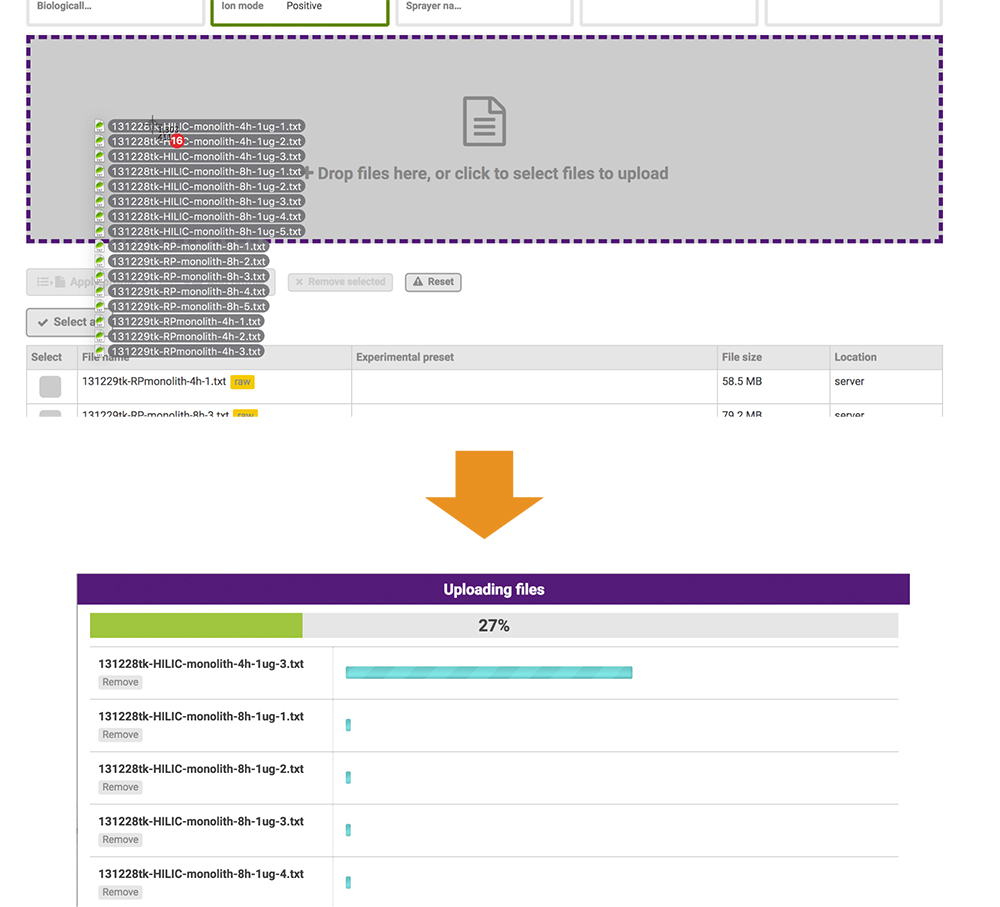
The files posted include raw data and peak lists from the mass spectromerter and identification results. Based on the PRESTO independently developed JavaScript library, the GlycoPOST file upload system enables users to upload files at higher-than-conventional speeds by expanding the standard functionality of the web browser (Fig. 19). This function allows all data-posting processes to be implemented via the web browser only.
Information on specific posting procedures and terminology is available at https://glycopost.glycosmos.org/help.