
Tamiko Ono
Tamiko Ono joined the JST Integration Promotion Program as a research assistant for the Kinoshita Lab in the Soka University Faculty of Science and Engineering in September 2017 and has been engaged in developing the GlyCosmos portal and collecting relevant data.

Kiyohiko Angata
After completing his doctoral studies at the University of Tsukuba Graduate School of Life and Environmental Sciences, Dr. Angata studied glycobiology at the La Jolla Cancer Research Foundation (presently the Sanford Burnham Prebys Medical Discovery Institute) under the tutelage of Professor Minoru Fukuda. In his current position at the National Institute of Advanced Industrial Science and Technology of Japan, he is conducting research to analyze glycan gene expression and new glycan functions, to find commercial applications of glycans, and to develop a glycan-related database (ACGG-DB).
In this seventh installment of this series, biological pathways involving glycoproteins, glycan-related genes relevant to disease, and a database on infectious diseases and glycans will be described. To understand the functional aspects of glycans, it would be useful to know which proteins are glycosylated, and the pathways involving glycoproteins (GlyCosmos Pathways). In addition, glycans are known to have a role in the pathogenesis, pathophysiology, and progression of a variety of diseases. This article also introduces diseases caused by mutations of genes involved in glycan synthesis and glycan degradation, and covers the features of databases on the relationships between infectious microorganisms and glycans (GDGDB and PACDB).
One of the most important issues in glycan research is the elucidation of the functions and roles of glycans. Previous installments in this series have introduced databases of structures of glycans, glycoconjugates, and molecules related to the biosynthesis and recognition of glycans. Of the glycoconjugates, glycoproteins in particular, are involved in a wide range of cellular functions, and a database, known as GlyCosmos Pathways, has been developed which enables searches for interactions with other molecules and outputs the results visually. Although there are other databases of binding molecules and pathways such as NCBI Gene and KEGG PATHWAY, GlyCosmos Pathways is distinguished by its focus on glycoproteins and the automated visual output that helps advance understanding of the localization and interaction of glycoproteins.
Genes involved in glycan biosynthesis (glycan-related genes, or glycogenes) include molecules relevant to biosynthesis such as sugar nucleotide synthases, sugar nucleotide transporters, and glycosyltransferases, as well as glycosidase genes, which are required for degradation. GDGDB compiles information about genetic diseases caused by mutations of these glycogenes, for example, congenital disorders of glycosylation (CDG) and congenital muscular dystrophy (CMD). OMIM (https://www.ncbi.nlm.nih.gov/omim, https://omim.org) provides detailed information about these diseases and knockout mice.
One of the key roles of glycans is that they serve as the “face of cells”. Glycans are involved in recognizing differences among cell populations forming tissues and in self-nonself discrimination, and many infectious pathogens recognize host-specific glycans: for example, influenza virus, which uses sialic acid on the cell surface as a receptor. PACDB has collected information about infectious pathogens targeting glycans expressed in human tissues, and provides information about the structures of glycans that bind each pathogen.
Using GlyCosmos Pathways, you can search for pathways that involve glycoproteins. In the example described herein, we extracted glycoprotein-related pathways from the Analysis Tools of the Reactome database 1, using UniProt 2 IDs annotated with glycosylation data.
Pathways can be searched by their species, pathway names, protein names, or gene symbols.
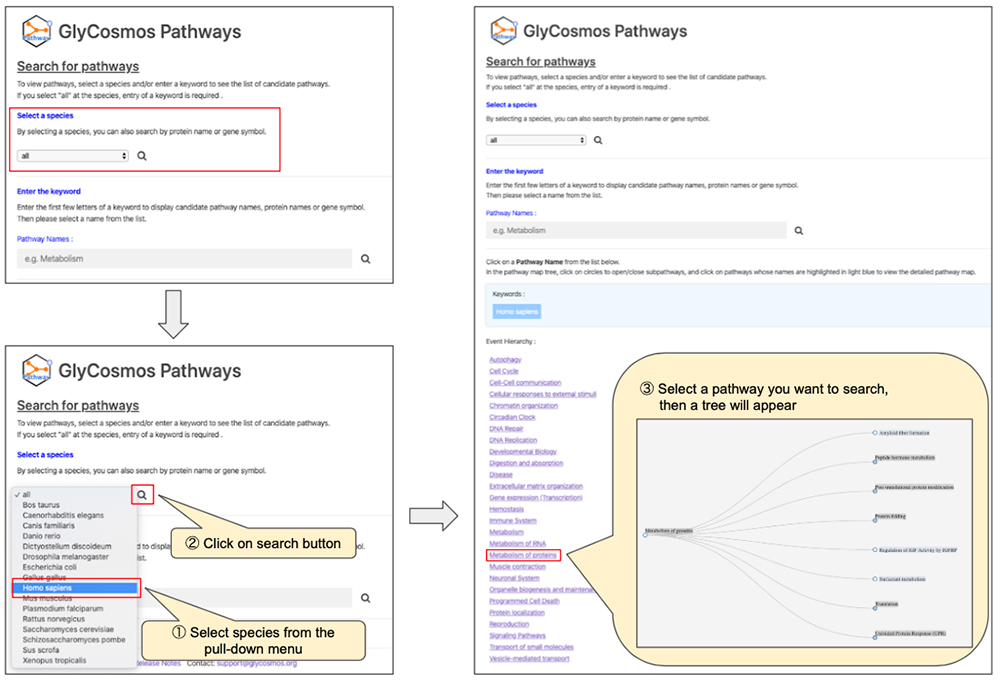
● Search by species (Figure 1)
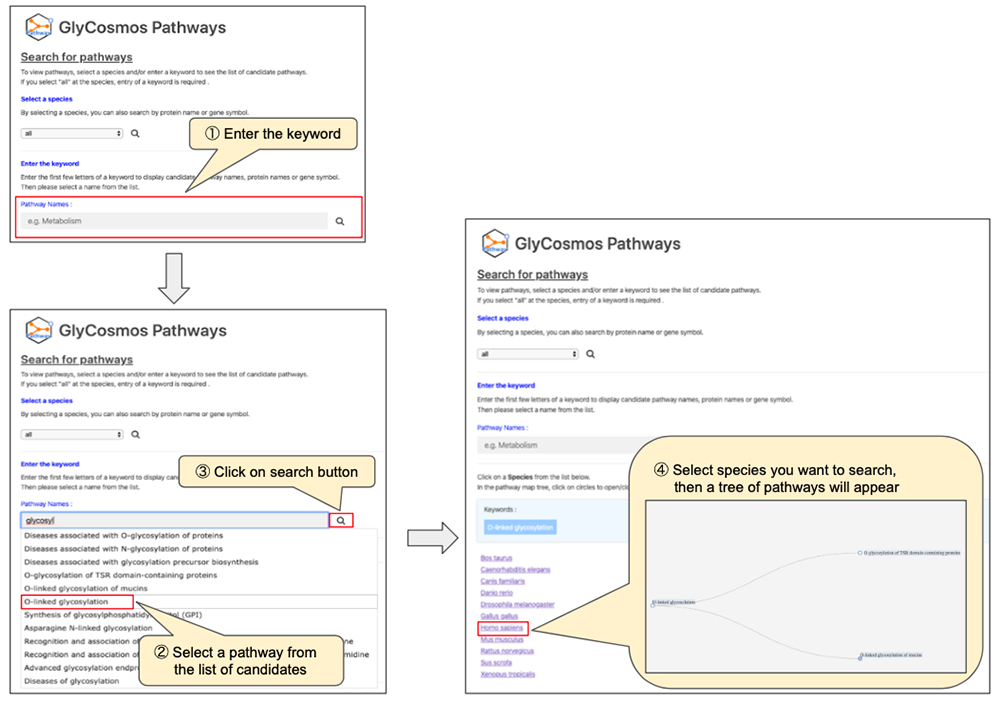
● Search by pathway name (Figure 2)
● Search by protein name or gene symbol (Figure 3)
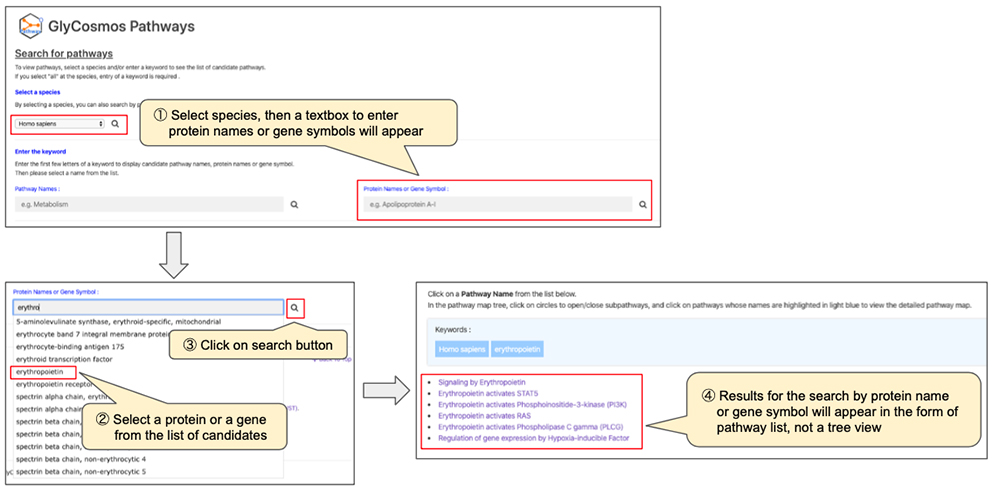
After narrowing down the list of pathways using the keyword search, look for the pathway of interest by following the tree (Figure 4). If you click on pathways shaded in gray, sub-pathways in the lower hierarchy would appear. If the pathway is shaded in light blue, a new window will open showing the page giving pathway details. When searching by protein name/gene symbol, trees will not appear; selection of the pathway will open a new window and show the page providing pathway details. You can obtain detailed information about the pathway in a new window (Figure 5).
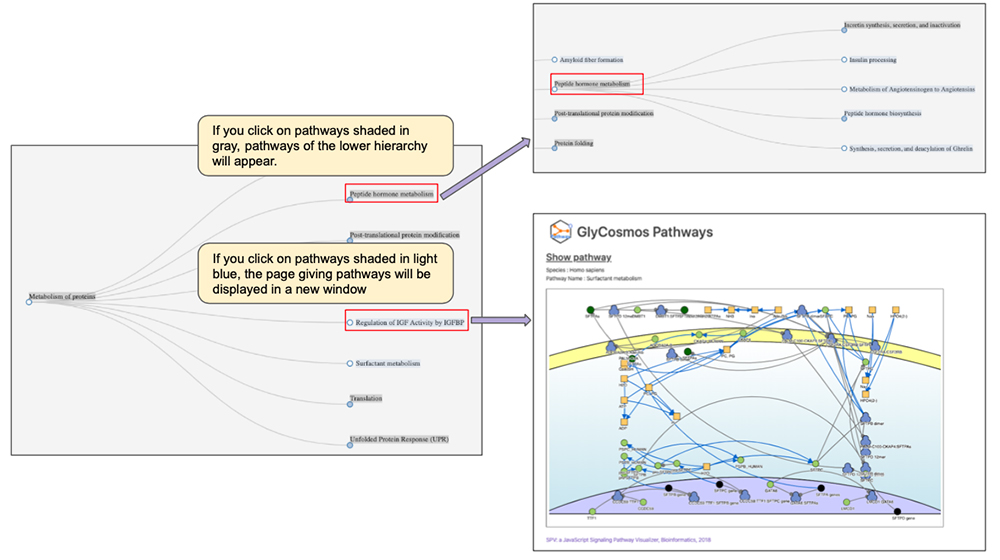
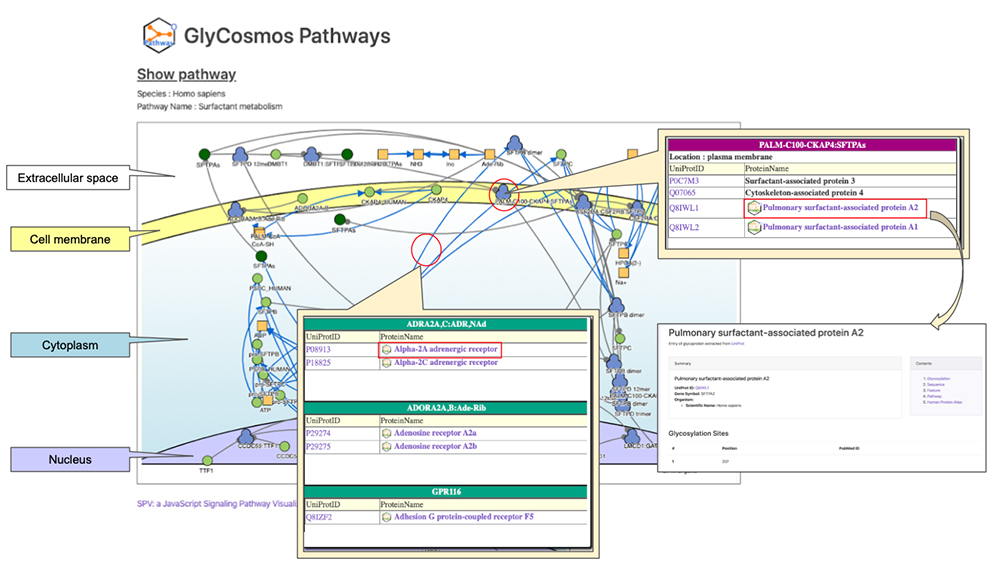
Pathways are visualized using the Signaling Pathway Visualizer (SPV) tool 3. Currently, for purposes of cellular localization, the cell is divided into four compartments: “extracellular space”, “cell membrane”, “cytoplasm”, and “nucleus”. We are further developing the tool so that cellular localization could also be shown on the organelle level in the future. If you click on each molecule, you can see the list of proteins relevant to the molecule and information about each protein including cellular localization, UniProt ID, and protein name. If you click on the arrows representing reactions, you can obtain information about catalysts, and if the catalyst is a protein, a list of UniProt IDs will appear. Glycoproteins are labeled with specific icons; if you click on an icon, you will move to the entry page of GlyCosmos Glycoproteins where you will get detailed information about the glycoprotein.
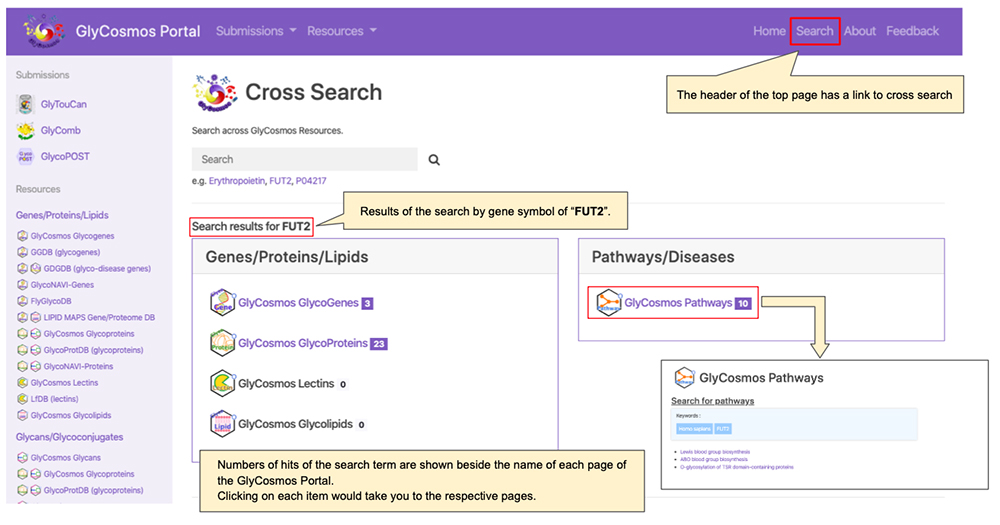
In the update of the GlyCosmos Portal in December of 2019, a cross search function (https://glycosmos.org/searches/cross_search) was newly added (Figure 6). From “Search” in the upper right part of the top page, you can move to the cross search screen. This function enables users to search glycogenes, glycoproteins, lectins, glycolipids, and pathways together at the same time, using only a single keyword. Items such as gene symbols, protein names, and UniProt IDs can be used as search terms.
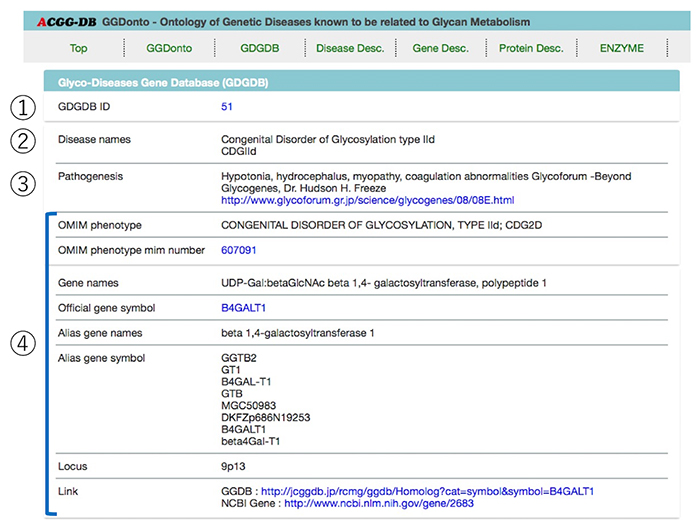
Approximately 300 human glycogenes have been cloned so far, and it has been revealed in recent years that some genetic diseases are caused by mutations in these glycogenes. Information about these diseases and mutations in the responsible genes is compiled in the GDGDB (Glyco-Disease Genes Database, https://acgg.asia/db/diseases/gdgdb, Solovieva et al. 2018 4). We herein introduce its features. *The GDGDB can be accessed not only from the ACGG website, but also from the websites of the Japan Consortium for Glycobiology and Glycotechnology (JCGG) and the GlyCosmos Portal.
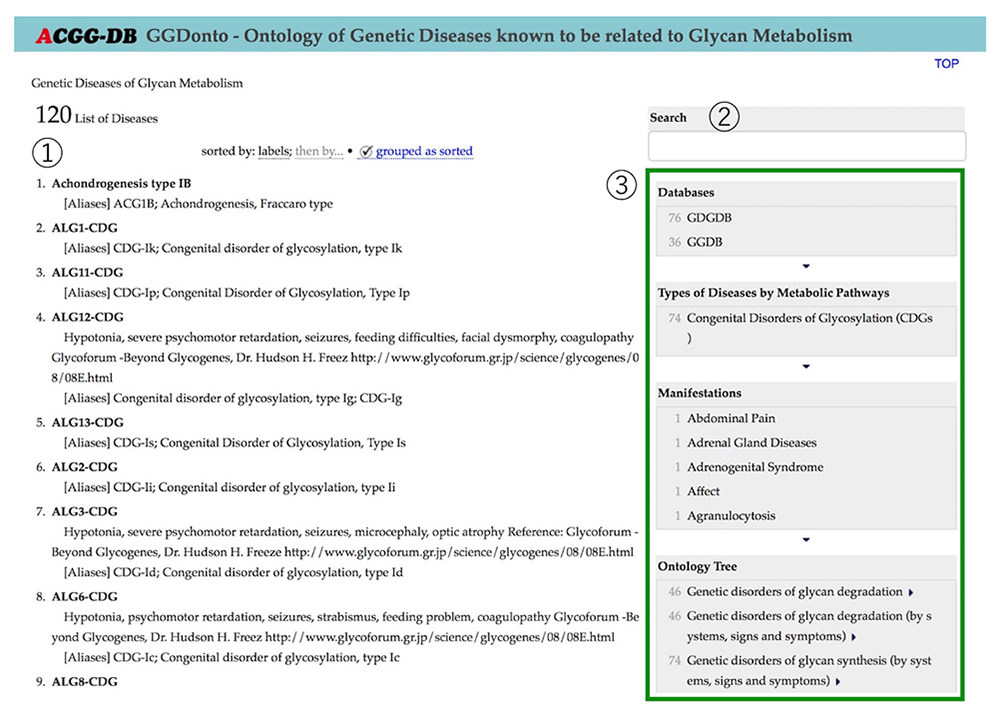
On the current top page of the GDGDB, 120 glycosylation disorders are listed in alphabetical order (Figure 7). By text search using keywords, or by a faceted search, you can narrow down the list of glycosylation disorders according to your purpose. Items selected for this narrowing down process include Databases, Types of Diseases by Metabolic Pathways, Manifestations, and Ontology Tree.
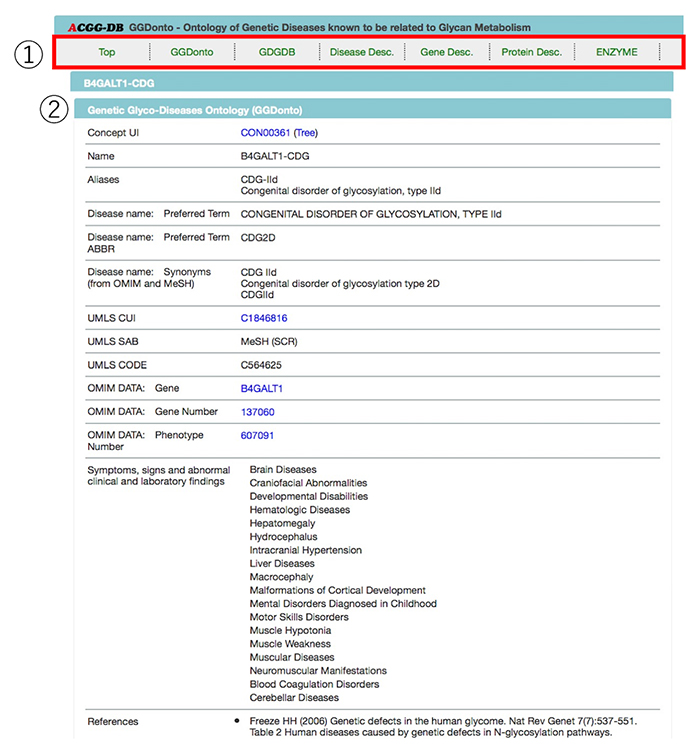
Selection of a disease name will take you to the details page. Summary of Genetic Glyco-Diseases Ontology (GGDonto) and GDGDB will appear on each selected glycosylation disorder page (Figure 8). Further, the page includes descriptions and links for information related to diseases (OMIM), glycogenes (GGDB and gene), proteins (UniProtKB), and enzymes (SwissProt/ENZYME), which can be used to collect more detailed information.
The summary of the GDGDB includes GDGDB ID, a description of the glycosylation disorder’s symptoms, some basic information about the responsible glycogenes and chromosomal location, and links to OMIM and the GGDB (Figure 9).
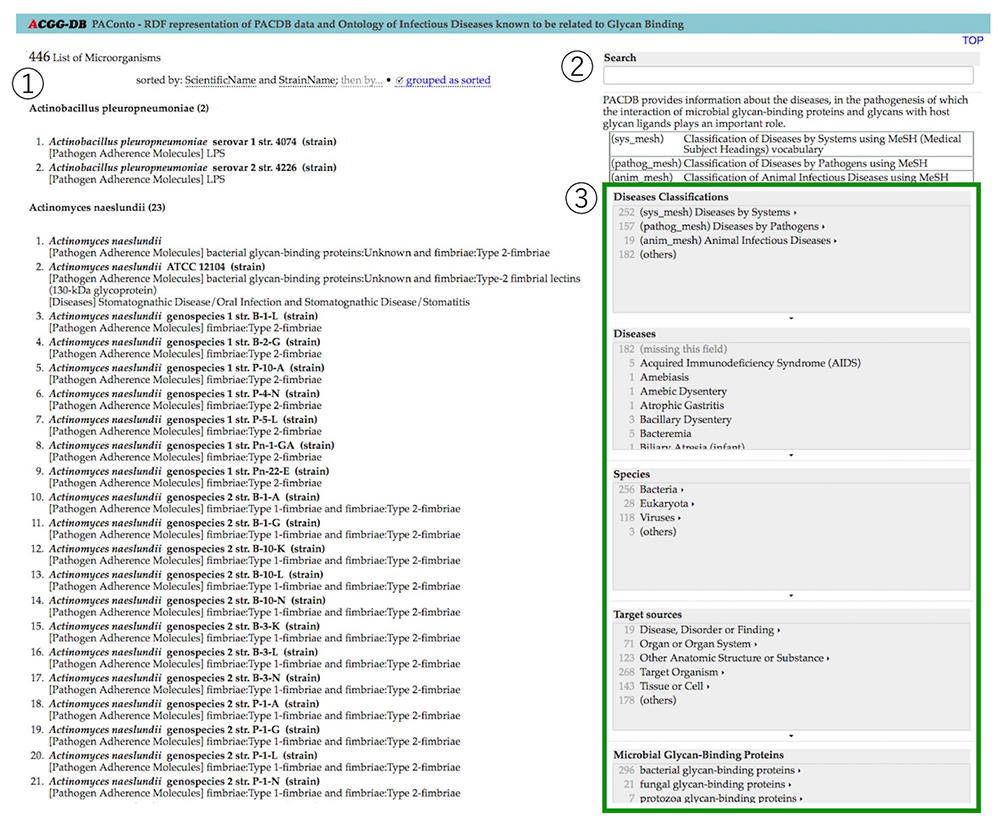
Many pathogens infect humans by binding to glycans on tissues. PACDB (Pathogen Adherence to Carbohydrate Database, https://acgg.asia/db/diseases/pacdb, Solovieva et al. 2017) has collected information on the glycan-binding profiles of bacteria, fungi, toxins, and viruses. Here we introduce the features of this database. *PACDB can also be accessed not only from the ACGG website, but also from the websites of the Japan Consortium for Glycobiology and Glycotechnology (JCGG) and GlyCosmos Portal.
On the top page of the PACDB, 446 microorganisms are listed in alphabetical order (Figure 10). By text search using keywords, or by a faceted search, you can narrow down the list of microorganisms according to your purpose. Items selected for this narrowing down process include Disease Classifications, Diseases, Species, Target Sources, Microbial Glycan-Binding Proteins, Pathogen Adherence Molecules Types, Glycans and Glycoconjugates Types, Monosaccharides, and Glycoepitopes, Structural Features of Carbohydrate Ligands.
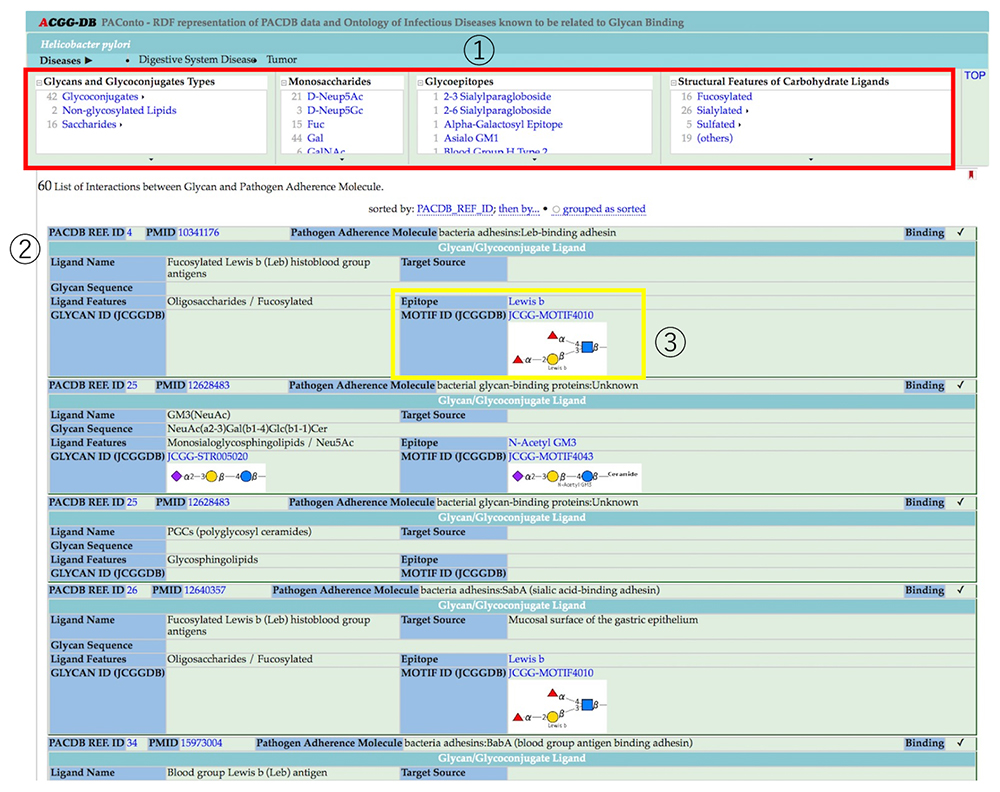
Selection of a microorganism name takes you to a page listing glycan structures to which the microorganism binds (Figure 11). Figure 11 gives the example of the Helicobacter pylori page, which lists 60 types of glycan-microorganism binding. By selecting the glycan structure (Glycans and Glycoconjugates Types, Monosaccharides, Glycoepitopes, Structural Features of Carbohydrate Ligands), the number of ligands can be narrowed down from the 60-ligand list. The PACDB displays models of glycan structures commonly listed in the ACGG-DB, which helps to visually understand the structures of glycans widely used as ligands by microorganisms. The page also provides information on epitopes or links to the JCGG-STR glycan structure database; you can check lectin affinity of binding to other receptors at the JCGG-STR site.
Recent studies have revealed that the glycans change upon malignant transformation, but the effect mostly remains to be elucidated. Whole-genome sequences of patients with a variety of genetic disorders are also being revealed, and more and more glycogenes responsible for these diseases are being identified. In addition, data on the ligands (glycans) and receptors (lectins) of infectious microorganisms and host cells continue to be accumulated. We are now trying to add data to the databases introduced in this article such as the GlyCosmos Pathways database, GDGDB, and PACDB, and develop cross-sectional use with other databases explained in this series.