
Hans-Joachim Gabius
Hans-Joachim Gabius obtained his MSc degree in 1980 and then his PhD in 1982 for chemical and biochemical studies on the proofreading mechanisms of aminoacyl-tRNA synthetases under the direction of F. Cramer, Max-Planck-Institute for Experimental Medicine, Göttingen, Germany. 1981 was mostly spent in the laboratory of J. Abelson at UC San Diego, investigating tRNA splicing. After starting work in tumor lectinology in 1983 at the Max-Planck-Institute in Göttingen, a post-doc period in the group of S. H. Barondes at UC San Diego (1984-1985) and appointments as associate professor for pharmaceutical chemistry in Marburg (1991) and as head of the Institute for Physiological Chemistry, Faculty of Veterinary Medicine, Ludwig-Maximilians-University Munich (1993) followed. He has edited several books on glycosciences, among them the textbook: The Sugar Code: Fundamentals of glycosciences, published in 2009. His track record comprises over 700 PubMed entries, close to 30,000 citations and an h-factor of 89.
Lectins have a central role in translating glycan-encoded information into bioactivity. They are classified according to the fold of their carbohydrate recognition domain (CRD). This structural unit is proposed to be capable to do (much) more than to interact with cognate glycan(s). As outlined for the proof-of-principle case of the galectin CRD, distinct sequence elements appear capable to extend a CRD’s functionality profile: they can implement molecular switches for activity, conformation and quaternary structure, even establish complementary regions for other types of binding partners, letting the CRD gain an unsuspected multifunctionality. By further testing and validating these suggestions, answers may be provided for major unresolved questions, e.g. why galectins belong to the small set of secreted proteins without a signal sequence. This case study teaches the salient lesson of the possibility for a CRD to consist of various structural elements beyond that binding the sugar. Of potential relevance for public health, the idea of a gene capture from the host as origin of viral adhesins that have a galectin-like fold (incl. coronaviruses such as SARS-CoV-2) may inspire innovative modalities to counter pandemic threats.
The concept of the genetic code has led to view biochemical oligo- and polymers as ‘messages’, ‘read’ by molecular complementarity (recognition processes). In this case, nucleotides constitute a coding platform (the first alphabet of life) that enables replication, copying of DNA into RNA and the biosynthesis of proteins. The letters of the second alphabet of life, i.e. amino acids, are their building blocks. Information ‘written’ in the sequence of nucleic acids (genes) is ‘translated’ into the sequence of proteins. The genetic code thus converts the language of nucleotide-based messages into that of the class of proteins.
Equally ubiquitous and even more abundant in mass, a third class of biomolecules (alphabet), i.e. complex carbohydrates 1, is found in Nature. The sugar alphabet is the molecular basis for letting the respective enzymatic machinery produce oligo- and polysaccharides. They are often attached as a glycan part to proteins and lipids so that cellular glycoconjugates are generated. Commonly found on the surfaces of cells and in the extracellular matrix, these linear and branched chains are readily accessible and can therefore readily engage in molecular interactions. Like nucleic acids or proteins, these biomolecules could embody a signal system that stores and conveys biological information, in this case based on sugars. Again, molecular complementarity will be involved in the flow of the information, here from the (sugar) code word to a recognition process that then triggers the actual cell biological effect.
Initially, as illuminated by the following verbatim quotations, “biological phenomena based on the specific recognition of sugar chains have [yet] attracted much less attention in comparison with those of nucleic acids and proteins” 2. “Only in recent years have we begun to appreciate how deeply glycan functions pervade all aspects of organismic biology, molecular biology, and biochemistry” 3, and the reader may wonder why this is the case, an answer phrased as follows: “In this remarkable age of genomics, proteomics, and functional proteomics, I am often asked by my colleagues why glycobiology has apparently lagged so far behind the other fields. The simple answer is that glycoconjugates are much more complex, variegated and difficult to study than proteins or nucleic acids” 4. Is such a high level of structural complexity a “whim of Nature” or the key to let glycans become “multipurpose tools” 5?
In fact, what at first sight indeed appeared mysterious, i.e. the multitude of cellular glycan structures (the glycome), as a phenotypic feature akin to a biochemical fingerprint 6-9, has turned out to receive a fundamental functional meaning by the concept of the sugar code: the exceptional chemical features of carbohydrates (together with the very sophisticated level of organization of the assembly lines for glycan synthesis in the Golgi apparatus, the structural equivalent of the ribosome in protein biosynthesis) enable to ‘write (oligosaccharide) messages’ with a minimum of letters that reach an optimum of biochemical coding capacity. In contrast to a peptide, there are more structural parameters available for glycan design than the sequence, that is the anomeric configuration (α/β) and the positions of the points of connection of each glycosidic linkage. Being small and diverse facilitates to present a plethora of signals with a different meaning on cell surfaces, a prerequisite to let glycans become multipurpose tools. Molecular complementarity between glycans and proteins accounts for the process to ‘select’ code words and to ‘read’ the glycan-encoded information (this activity is the etymological root of the term ‘lectin’, which was derived from the Latin verb legere 10), this recognition underlying cellular activities such as adhesion or proliferation 11-14.
Nowadays, the definition of a ‘lectin’ automatically implies the respective protein to bind glycan(s), and this by a module called the carbohydrate recognition domain (CRD) 15. Of physiological significance, a puzzle-like combination of such a structural unit with other types of modules to form hybrids (multimodular proteins) is commonly seen in families of lectins such as C-type lectins, galectins or siglecs. It opens a door to functional versatility, e.g. by building the long molecular tentacles (stalks) characteristic for potent cell adhesion molecules acting in inflammation (selectins) or by using collagen-like repeats as molecular glue for the reversible self-association known from collectins, ficolins or galectin-3 (Gal-3) 2,16-19.
The term ‘CRD’ is structurally defined by the nature of the fold that establishes the carbohydrate-binding region. Of note, it is often interpreted not only as a structural unit but also as a functional entity. By exclusively considering a CRD as receptor for its cognate glycans, this term acquires a dogmatic character. It would mean missing opportunities in the quest of reaching a full understanding of (ga)lectin functionality when setting such an inherent strict limitation to our interpretation of what a CRD is capable to do. Here, the idea is to put forward that instead such a domain is a versatile platform that integrates various biorelevant residues/regions in an intra-CRD puzzle.
Explicitly, the CRD (of a (ga)lectin) is proposed i) to consist of (much) more than the contact site for glycan ligands and ii) to be able to do (much) more than to bind a glycan. Looking at galectins as instructive proof-of-principle example, this CRD is introduced, then first evidence is presented for occurrence of molecular switches within the CRD and finally examples of its emerging diversity to ‘select’ and ‘read’ are given. By making contacts to proteins, even simultaneous binding to two types of counterreceptors (glycan and protein) and teamwork of these sites are possible.
The structural platform of the family of galectins is the β-sandwich (jelly roll) motif with two anti-parallel β-sheets of six (S1-S6) and five (F1-F5) β-strands 20,21 As exemplarily shown in Fig. 1A, two such units can associate to form a homodimer with cross-linking capacity, a hallmark of (proto-type) galectin-dependent lattice formation and biosignaling 22 A set of seven conserved amino acids, called the sequence signature, has not only been the common denominator for allowing classification but is also responsible for shaping molecular complementarity with the canonical ligand lactose, and their spatial presentation in the binding site is mostly preorganized like a lock for a sugar key (Fig. 1B,C). Strategically, its central Trp residue enables C-H/π bonding with the B face of galactose of the disaccharide lactose, and hydrogen bonding to the axial 4’-hydroxyl group and the exocyclic CH2OH (at C6’) of the galactose moiety ensures specific recognition of this epimer (excluding mannose and glucose) without a substitution at C6’ (thus, α2,6-sialylation will mask a galactose residue). Sequence variations around these (mostly) conserved amino acids seen among the family members establish the individual features of the architecture of the carbohydrate-binding region. Systematic exploration of structure-affinity relationships has characterized the glycan-binding profiles for the members of this lectin family, for example by using frontal affinity chromatography 23, that result from the mentioned sequence changes. Central to paving the way toward purification, structural analysis, then sequence comparisons and specificity assays was the observation that presence of a reducing agent (dithiothreitol) was essential to preserve β-galactoside-dependent haemagglutination activity in extracts of tissues, first described for organs of the electric eel and rat and found to be especially strong in chicken embryo (pectoral muscle) 24-26. This pioneering work gave reason to trace the first molecular switch in the galectin CRD based on a chemical change.
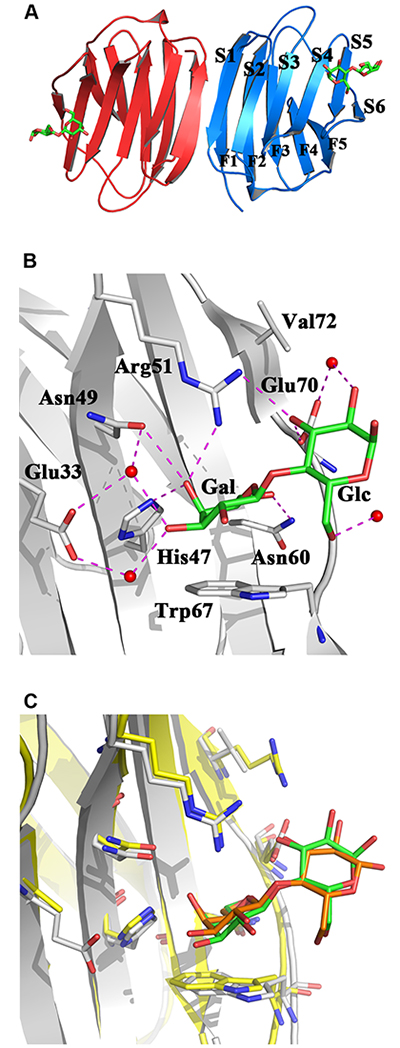
When searching for answers to the question on the molecular basis of the loss of activity as haemagglutinin (aggregating erythrocytes by letting a galectin bridge them) by oxidation, indications for existence of more than a single explanation have been found. In the first galectin that had been purified, i.e. electrolectin (from the electric eel, Electrophorus electricus) which has no Cys residue, oxidation of the central Trp to oxindole has been suggested to cause this galectin’s vulnerability to oxygen 27. Interestingly, oxindole generation and serious harm to the β-galactoside-binding activity are not common among galectins, the connection between these two processes a still unresolved issue. In mammalian and avian proto-type (homodimeric) galectins, their Cys residues appear to come into play, and this probably in different manners: intramolecular disulfide bridging and oxidation of sulfhydryl to sulfenate or sulfinate, even to sulfonate are being discussed for Gal-1 and -2 28-31. However, mechanistic details of the way these proposed altered chemistry-based molecular switches work, for example how rather distant sulfhydryls, especially Cys2/Cys130 of Gal-1, come together in a disulfide bridge and how this triggers the dissociation of the homodimer and loss of lactose binding, are not yet perfectly clear. What truly is remarkable is that involving Cys residues can most likely operate in a specific manner among galectins; what is clear is that presence of these moieties is not required for carbohydrate binding of human Gal-1 32,33, as also noted for the Cys-less electrolectin above. In this respect inspiring, the analogy to an alarmin such as the high-mobility group box 1 (HMGB1) protein (and, notably, galectins play roles in innate immunity after their non-conventional secretion like alarmins 34,35) tells us something of great relevance: different oxidation states of the sulfhydryl “license three mutually exclusive functions: alarmin, chemoattraction or tolerance” 36. And there is more to add already: as the case of the chicken galectin (CG)-1B teaches, cystine generation is possible within a CRD (Cys2-Cys7) and between CRDs (Cys7-Cys7’), leading to substantial shape changes 30. Clearly, Cys residues deserve renewed interest to elucidate their nature as switches of lectin activity, quaternary structure and global shape.
In addition to Cys residues, another type of molecular switch based on a chemical aspect of an amino acid has very recently been detected: the assignment work of NMR-spectroscopical signals on human Gal-7 revealed presence of two conformers, whose origin has been attributed to the cis/trans-isomerization of the peptide bond toward Pro4 37. At present, it is an open question as to whether this type of switch (i.e. the cis-trans isomerization) may affect binding properties for cognate oligosaccharides (or other types of ligands; please see below). In such a case, it would work like an allosteric effector, which modulates affinity for binding partners by shifting the conformer equilibrium. Talking about allostery for a galectin, first evidence for presence of such a binding site has been reported for mannose and congerin P (the third galectin from the Japanese conger eel, Conger myriaster) 38 and for a synthetic compound (a calix[4]arene) and human Gal-3 39, another feature of the CRD beyond binding a glycan ligand. Interestingly, the site of contact for the latter modulator was assigned to the galectin’s F-face, spatially separate from the β-galactoside-binding S-face 39. Considering the emphasis given to glycan binding, this region has so far not received that much attention, although it is in principle a docking site. Equally important, sequence diversification among galectins concerns also the noted sequence signature (please see information on the seven amino acids in crystal structures presented in a Fig. 1B,C and in the alignment shown in a Fig. 2A). Compensating a loss of β-galactoside-specific binding, a new receptor characteristic may arise in the CRD. Indeed, galectins are now gaining the status of proteins with specificity to more than one class of biomolecules. Toward this end, they can involve these two parts of the CRD, which can even cooperate with each other, and the contact site for lactose can be converted to a region complementary to a protein counterreceptor.
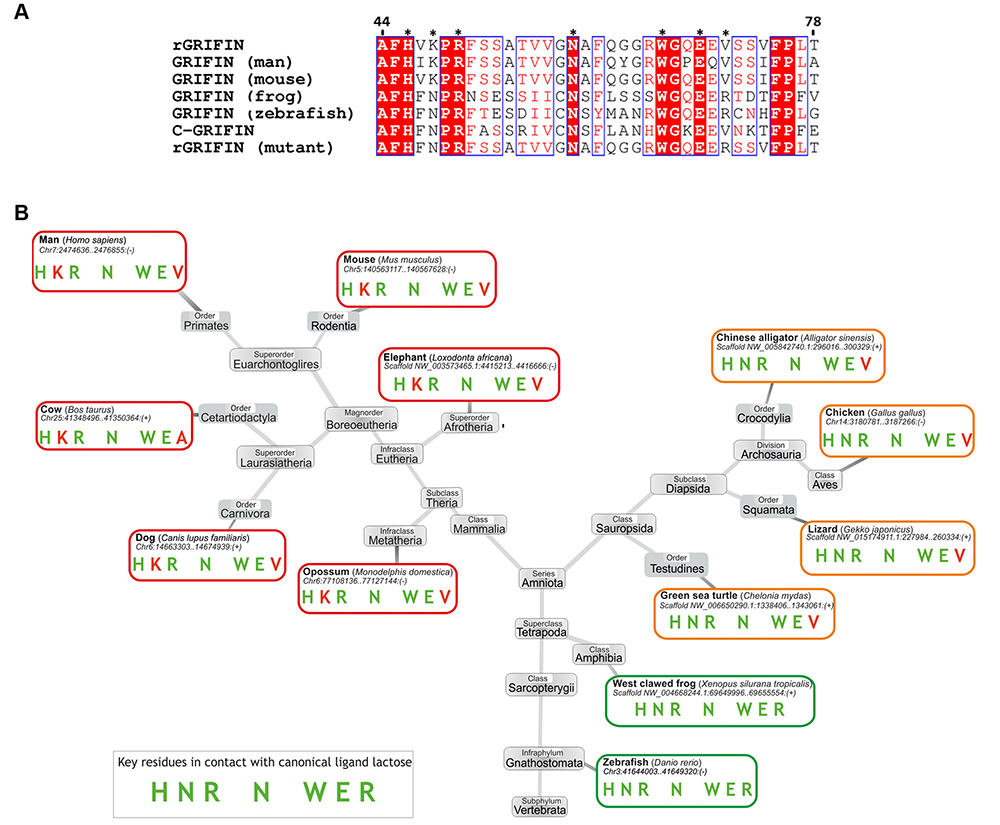
The analysis of binding of proteins by galectins started with the observation that Gal-3 confers resistance to apoptosis induction to human leukemic T cells. The detection of an anti-apoptotic activity and of sequence similarity between this galectin, in particular the NWGR motif, and members of the Bcl-2 family (involved in suppression of apoptosis) led to the demonstration of a lactose-inhibitable contact between Bcl-2 and Gal-3 40,41. The description of more cases of specific galectin-protein pairing followed, some already structurally defined in detail such as complex building of Gal-3 with the chemokine CXCL12 in solution 42 or of the C-terminal CRD of Gal-8 with the autophagy receptor NDP52 in crystals 43,44, others such as the association of the eye lens galectin (GRIFIN) with crystallins to contribute to forming the ‘biological glass’ of the eye lens 45 waiting for study on the atomic level (please note: this galectin presents deviations from the sequence signature in mammals but not birds as shown in Fig. 2). Obviously, the given names of interaction partners and of processes such as autophagy, immune regulation or tight protein packing signal a fundamental dimension of galectin functions, and the next paragraph introduces the amazing role of galectins as intracellular guardians of membrane integrity.
Intriguingly, their biosynthesis in the cytoplasm predestines galectins to serve in surveillance of endomembrane integrity: sensing damage to vesicles in the cell by the exposure of glycan chains of otherwise intraluminal (hidden) glycoproteins, associating to them and calling for help, that is then acting as bridge and forming a complex with components of the autophagy/repair machineries (as for example with the mentioned NDP52) strategically teams up binding of galectin to glycan and to protein counterreceptors 46,47. Extracellularly, chemokine binding to the F-face and glycan binding to S-face of Gal-3 gives a precedent for a bridging of two types of binding partners via two sites on the CRD of Gal-3 42. Studying the recognition of protein (and also glycan) counterreceptors intra- and extracellularly can thus become a fertile area.
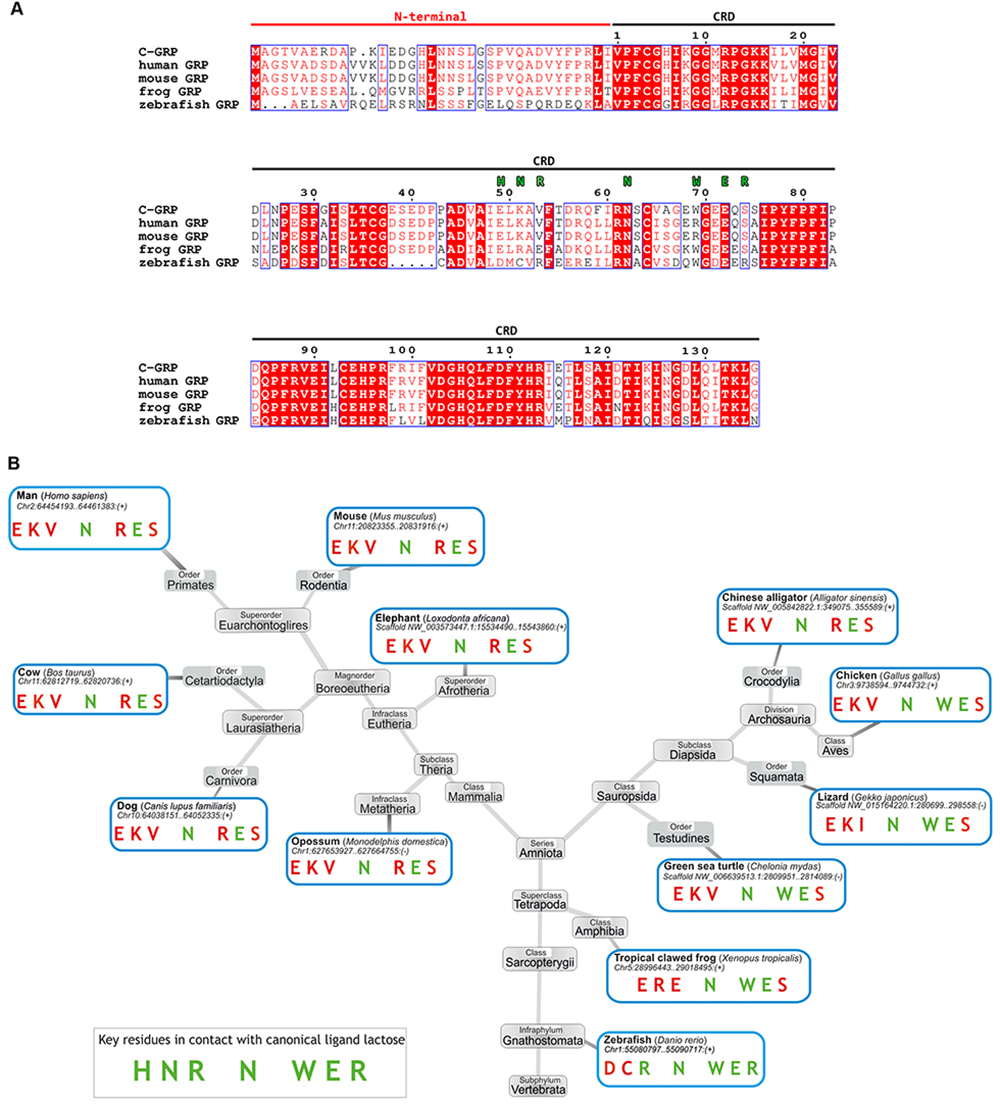
In this context, the findings of occurrence of sequence deviations in the (so-called) signature sequence for new members of the galectin family can broaden the range of biofunctionality of the galectin CRD 48. Interestingly, the Trp-to-Arg substitution has been elucidated to cause loss of lactose binding, instead installing preference for chitooligosaccharides and di-N-acetylated lactose (LacdiNAc) in a mushroom galectin, i.e. Coprinopsis cinerea galectin-related lectin 3 49. Such a loss and an acquisition of a new binding capacity (for a GalNAc) are caused by a cis-trans isomerization at Pro45 in a mushroom (Agrocybe cylindracea) galectin named ACG 50,51. The Trp-to-Arg substitution is seen to occur in vertebrates in the galectin-related protein (GRP): a respective phylogenetic is provided in Fig. 3 52. What its manifestation means for this member of the family has not yet been clarified. Of note, the exceptionally high degree of sequence conservation among GRPs from various species “suggests the entire coding sequence is under very strong positive selection, as generally seen for genes encoding proteins with multiple aspects involved in critical interactions, for instance with self, other proteins, and specific ligands” 48, a statement that underscores enormous interest to understand the reason for the positive selection and to shed light on the nature of the actual interactions with this special CRD.
The same applies to the potential of research that is based on the detection of the galectin fold in viral and protozoan proteins. This had been done by crystallographical analysis of the N-terminal fragment (VP8*) of the rotavirus outer capsid spike protein VP4 (53). This modular receptor for viral adhesion to host cells and the subsequent membrane penetration was proposed to have arisen “by the insertion of a host-derived, galectin-like CRD into an ancestral membrane interaction protein” 53. The study of further viral adhesins led to the following discoveries: sequence variability at maintained β-sandwich (galectin-like) fold design and binding properties to either glycan or protein parts of distinct host (glyco)proteins were, of note, observed also for spike proteins of various coronaviruses, shaping the hypothesis of a recruitment of the galectin fold as structural unit for this type of adhesin 54, and this is apparently not a unique case among CRDs. Fold similarity of adhesins/invasins of parasitic bacteria and viruses had been disclosed in the case of the CRD of a second large class of mammalian lectins, i.e. C-type lectins: “these are involved in interactions with the animal host and are either hijacked host proteins or their imitation” 55. The given hypothesis suggests that to counter viral threats one could take advantage of experience from working with galectins to test approaches to interfere with the contact between the (corona)viral (galectin-like) adhesin and its cellular counterreceptors 56.
The structural characterization of the ‘readers’ of sugar-encoded information (lectins) has uncovered a panel of folds (for galleries of respective illustrations, please see 57-59). By definition, their common property is the binding of glycan ligands. As seen for galectins and other types of sugar receptors such as C-type lectins 23,55 or lectins with the mannose-6-phosphate receptor (P-type) homology (MRH) domain 60, the specificity to glycans can cover a broad range in a family established by gene duplication and subsequent divergence. The sequence diversification can also turn a glycan- to a protein-binding motif. In addition, this property can develop at other sites of a CRD, as shown for the F-face of galectins, so that the resulting bifunctional protein acquires the attractive potential of the two sites for protein and glycan binding to engage in productive teamwork. In addition, molecular switches, e.g. by Cys oxidation or a cis/trans prolyl peptide bond isomerization, appear to be able to affect binding properties. The exemplary documentation of such cases for galectins is given with the intention to sensitize readers for realizing possibilities of CRD activity beyond carbohydrate binding and for exploring this hypothesis. In this sense, the presented informations give direction for research, that is to examine the concept for occurrence of altered chemistry-based molecular switches, to elucidate their molecular nature and mode of operation systematically, to define the complete contact and counterreceptor profiles for CRDs and to characterize the importance of the proposed puzzle-like intra-CRD architecture for the activity of each galectin, to gain a comprehensive level of understanding. Revealing their intra- and extracellular missions, the current status of research succinctly summarized recently 61, and the molecular key to make specific choices 62 continues to be an exciting and highly rewarding challenge. And answering such “Unsolved Questions about Galectins” will be sure to be recognized as a milestone along the route of “epoch-making events in the history of galectin research” 63
Acknowledgements and Note
Inspiring discussions with Drs. B. Friday, A. Leddoz and A.W. L. Nose are gratefully acknowledged. An apology is offered for the less than comprehensive coverage of the literature (in order to focus on outlining the concept and its perspectives).