
Ryuichiro Suzuki
Assistant professor at Akita Prefectural University since 2012. He studied the rational engineering of xylanase under Prof. Tsunemi Hasegawa and Dr. Atsushi Kuno in the Graduate School of Science and Engineering, Yamagata University. After graduating from Yamagata University (Ph.D, Science) in 2005, he studied the structure and function of R-type lectins under Dr. Jun Hirabayashi at the National Institute of Advanced Industrial Science and Technology. In 2006, he moved to the laboratory of Prof. Shinya Fushinobu (University of Tokyo) and studied the structure and function of enzymes involved in the metabolism of milk oligosaccharides (2006–2010). He then moved to the laboratory of Dr. Kazumi Funane at the National Food Research Institute, where he studied the structure-based design of cyclic isomaltooligosaccharides metabolism-related enzymes (2010–2012). At present he is assistant professor in Akita Prefectural University with Prof. Eiji Suzuki (2012–). His research interests are structure-function relationships of enzymes related to α-glucan metabolism.

Eiji Suzuki
Professor at Akita Prefectural University since 2011. After graduation from the Department of Biochemistry, Faculty of Science, Saitama University in 1985, he received his Ph. D. from the Graduate School of Science, University of Tokyo in 1990 under the supervision of Prof. Shigetoh Miyachi. He started his professional career in the Department of Biology, Ibaraki University as Assistant Professor (1990–1997) and then Associate Professor (1997–2001) in the Department of Environmental Sciences. He then moved to Akita Prefectural University as Associate Professor (2001–2011). His research interests are physiology, diversity, and evolution of carbohydrate metabolism in photosynthetic organisms, especially cyanobacteria.
Starch, a polysaccharide composed of glucose molecules, is formed during photosynthesis by Archaeplastida and is found in staple food crops such as cereals, tubers, and beans. Starch is utilized not just for food and food additive purposes but also for industrial purposes, e.g., in the manufacture of glues and bioplastics. Starch consists of amylose and amylopectin. The primary constituent amylopectin has an orderly branched structure and its structure has an effect on the taste and physicochemical properties of starch. Starch synthase, branching enzyme, and debranching enzyme are involved in amylopectin biosynthesis, but the mechanism controlling the assembly of highly ordered structures remains to be elucidated. Understanding the mechanisms controlling amylopectin’s structure opens up the possibility of designing an amylopectin with a favorable structure and properties. The authors have clarified the mechanism controlling the length of branched chains formed by branching enzymes. In this article, we describe the problems that need to be solved in order to control the structure and properties of amylopectin and recent progress in studies on branching enzymes. The production of structure-controlled amylopectins contributes to the development of our nation by raising the level of its food self-sufficiency and helping it reach the goal of a decarbonized society.
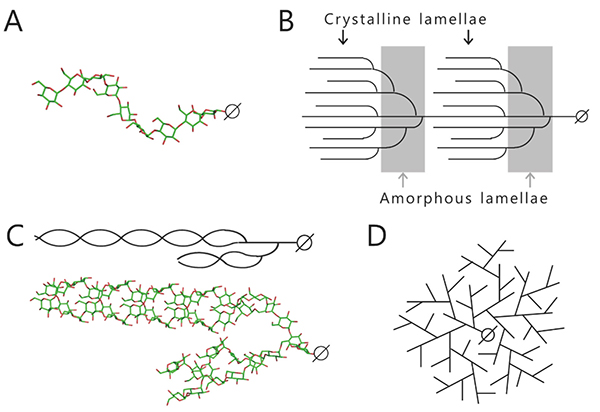
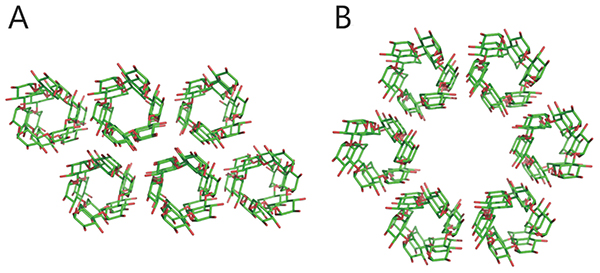
Starch and glycogen are known as storage polysaccharides (branched α-glucans) and their structures and properties differ from each other. Starch consists mainly of amylopectin (approximately 65–85%) and amylose (15–35%). Amylose is an essentially linear polymer of 500–20,000 α-1,4-linked glucose units1. The linear α-1,4-linked glucan chain of amylose forms a helix via intramolecular hydrogen bonds, although it contains a few α-1,6 branch points (less than 1%)1,2 (Fig. 1A). Amylopectin is an orderly branched polymer made up of 10,000–100,000 α-1,4-linked glucose units with 5%–6% α-1,6 branch points1. Amylopectin has a tandem-cluster structure in which multiple grape-like clusters are tandemly linked1,3,4 (Fig. 1B). The tandem-cluster structure is an assemblage of repeating highly and scarcely branched regions (Fig. 1B), and consequently neighboring glucan chains in scarcely branched regions form a double helix structure (Fig. 1C). Formation of the helix structure with elimination of water molecules is needed for the polymers to show crystallinity and become water-insoluble. The highly and scarcely branched regions have been designated amorphous lamellae and crystalline lamellae, respectively1,3,4. The crystalline lamellae of starches extracted from cereals and tubers are known to have A- and B-type crystal packing arrangements, respectively1 (Fig. 2). A C-type crystal packing arrangement, a mixture of A- and B-type crystal packing, is also known1.
Amylose and amylopectin can be stained by forming colored inclusion complexes with iodine. Amylose consisting of long glucan chains is stained violet blue, while amylopectin having short glucan chains is stained red violet.
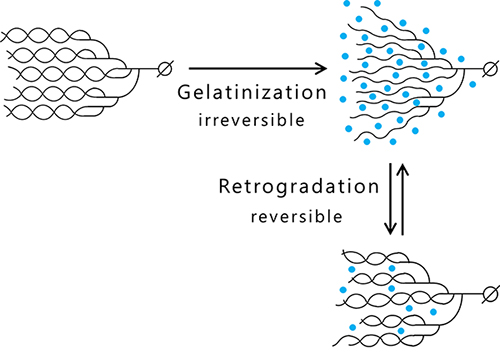
Amylose and amylopectin are gelatinized by heat-moisture treatment that causes unwinding the helix structure due to water molecule incorporation into the clusters and results in the acquisition of viscosity as a material property. Cooling of the gelatinized glucan induces retrogradation, which is accomplished by exclusion of many water molecules and restoration of the helix structure, and the viscosity is lost4 (Fig. 3). Amylose easily undergoes syneresis and retrogradation because it is composed of long glucan chains. In contrast, amylopectin having much shorter branched chains resists retrogradation.
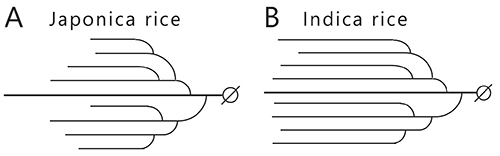
The effect of amylopectin and amylose behaviors on the tastes of cooked (steamed) rice is explained simply below. The rice cultivars, indica and japonica, are the most commonly crop varieties. The texture of steamed japonica rice is sticky and fluffy, whereas that of steamed indica rice is dry. The difference in the texture can be explained by the amylose content of the endosperm starch, which is about 20% for japonica rice and about 25% for indica rice. The hardness or softness of the steamed rice is correlated with amylose content4 and the dry texture of steamed indica rice is due to the high amylose content. The amylopectin branched chains are shorter in the endosperm of japonica rice than indica rice (Fig. 4). This is attributed to the starch synthase (SS) isoform IIa (SSIIa) that catalyzes elongation of the branched chains, which has lower activity in japonica rice than indica rice3,4 (see “5. Enzymes involved in amylopectin and glycogen biosynthesis”). The structure of amylopectin is also related to the texture of steamed rice, since retrogradation of short and long branched chains in amylopectin progresses slowly and quickly, respectively.
Glycogen is a randomly branched polymer consisting of linear α-1,4-linked glucan chain and α-1,6 branch points. Water-soluble amorphous glycogen is highly branched (contains 8–10% α-1,6 branch points) and has short branched chains (see “3. Method of structural analysis of branched α-glucan”)1 (Fig. 1D). Linear glycogen chains cannot form helix structures and consequently they are hardly stained at all by iodine.
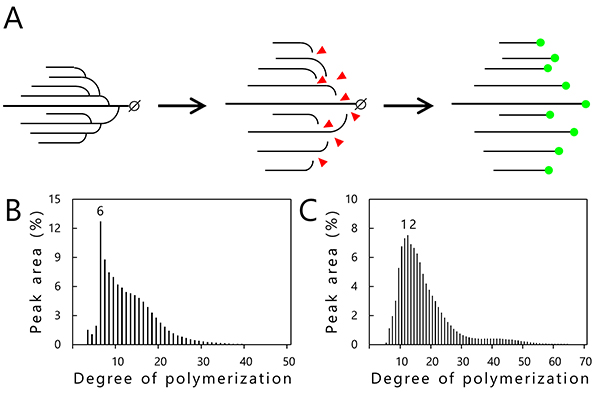
Chain-length distribution analysis by capillary electrophoresis is used to investigate the structure of amylopectin and glycogen5. In this analysis, branch points (α-1,6-linkage) of the α-glucans are specifically hydrolyzed using a debranching enzyme (isoamylase) to obtain linear α-1,4-linked glucan chains (Fig. 5A). Reducing ends generated by the debranching treatment are labeled by a fluorescent dye (APTS; 8-amino-1,3,6-pyrenetrisulfonic acid) (Fig. 5A) and separated by capillary electrophoresis. The resultant data representing the relationship between the degree of polymerization (DP) of each glucan chain (horizontal axis) and percentage of glucan chains of different length (vertical axis) are shown graphically in Figs. 5B and 5C5. Commercially available glycogen from oyster primarily consists of branched chains with DP6 (Fig. 5B). On the other hand, purified amylopectin from rice endosperm (japonica variety Nipponbare) has a peak with glucan chains of DP10–13 (Fig. 5C), indicating that the clusters of amylopectin contain branched chains mainly of 10 to 13 glucose units. The chain-length distribution analysis for determining the length and number of the branched chains is a powerful structural analysis tool. However, the location of the branch points and mechanism of helix structure formation (Fig. 1C) are not provided, restricting us to two-dimensional model predictions exclusively3 (Fig. 1B). It is difficult to determine the structure of the branched α-glucans even though they consist only of glucose molecules.
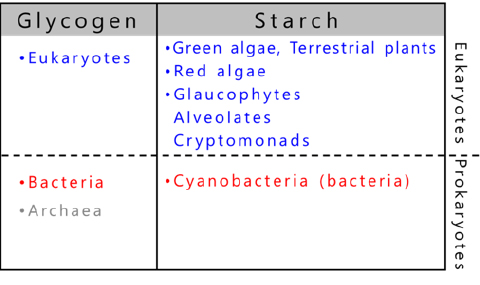
The organisms that store glycogen are widespread in nature and use glycogen as an energy source6,7 (Fig. 6). In contrast, starch is a photosynthetic product of a limited number of eukaryotes6-9. The starch-producing organisms include green algae, terrestrial plants evolved from green algae, red algae, glaucophytes as well as cryptophytes and alveolata derived from red algae9 (Fig. 6). Most of the several thousand species of cyanobacteria10 are photoautotrophic prokaryotes and generally produce glycogen as storage α-glucan, but about 10 species exceptionally accumulate amylopectin (referred to as cyanobacterial starch)8,11 (see “5. Enzymes involved in amylopectin and glycogen biosynthesis”).
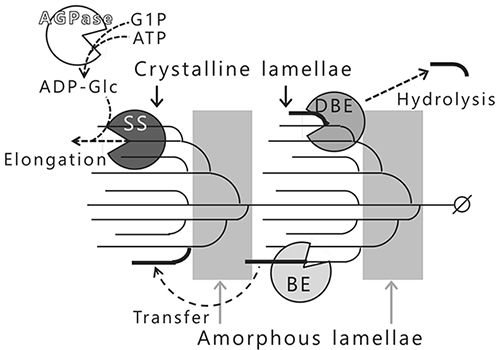
Amylopectin biosynthesis is carried out by at least four classes of enzymes3,4 (Fig. 7). ADP-glucose pyrophosphorylase (AGPase; EC 2.7.7.27) catalyzes the production of ADP-glucose (ADP-Glc) from glucose-1-phosphate and ATP, and the resulting ADP-Glc is used as a donor substrate for starch synthase (SS; EC 2.4.1.21). SS catalyzes the transfer of glucosyl moiety from ADP-Glc to the non-reducing end of a growing glucan chain via an α-1,4-linkage. The branched chains in amylopectin are elongated by repeated reactions of SS (Fig. 7). Branching enzyme (BE; EC 2.4.1.18) catalyzes the intra- or intermolecular transglucosylation to form new branch points (α-1,6-linkage) by cleaving the α-1,4-linked glucan chains that have been elongated by the SS to no less than DP12 (Fig. 7). BE infrequently forms improper branch points in the crystalline lamellae. The improper branch points (α-1,6-linkages) are specifically hydrolyzed by the debranching enzyme (DBE, isoamylase [ISA; EC 3.2.1.68]) forcing amylopectin to adopt a crystalline structure (Fig. 7). Amylopectin in terrestrial plants is synthesized by SS, BE, and DBE activities in the right positions3,4,12.
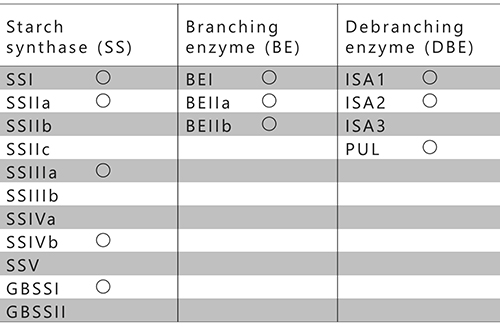
The currently known SS, BE, and DBE isozymes from rice are summarized in Table 1. Rice has eleven SSs, three BEs, and four DBEs and these isozymes are involved in the metabolism of assimilatory or storage starches13 (Table 1). Among the four DBEs, ISA1 and ISA2 participate in the amylopectin biosynthesis14,15, while ISA316 and pullulanase (PUL)17 are involved in degradation of the amylopectin. However, PUL has been found to partly participate in amylopectin biosynthesis17. The participation of AGPase, SS, BE, and ISA in amylopectin biosynthesis is common in algae and terrestrial plants.
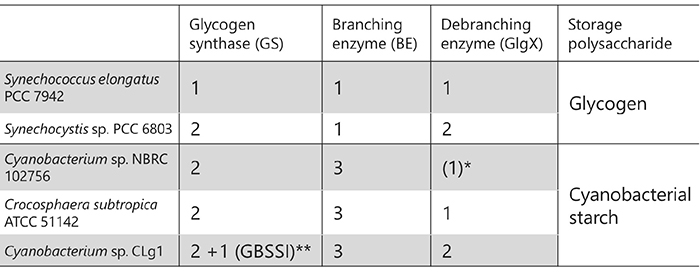
Glycogen biosynthesis-related enzymes in bacteria are homologous to the enzymes involved in amylopectin biosynthesis and they share high amino acid sequence identity6,18. However, SS corresponds to glycogen synthase (GS), and DBE corresponds to ISA or GlgX in bacteria19. GS and BE are involved in glycogen biosynthesis, but DBE in bacteria unlike that in amylopectin-producing plants only participates in glycogen degradation. The number of related isozymes is smaller in glycogen-accumulating organisms than in plants. For example, Escherichia coli has only one each of GS, BE, and GlgX. Although they are prokaryotes, cyanobacteria are unusual in that some of them are amylopectin-accumulating species8,11 (see “4. Distribution of organisms storing glycogen or starch”). The amylopectin-accumulating cyanobacteria have fewer related isozymes compared to plants8,20-22 (Table 2). In addition, there is currently no evidence that cyanobacterial enzymes form the protein-protein complexes reported in terrestrial plants (see “6. Effects of biosynthesis-related enzymes on the structure and properties of starch”). These facts suggest that cyanobacteria have the simplest and most basic form of the amylopectin biosynthesis system on earth. Additionally, it is of interest that amylopectin-producing cyanobacteria have a lot more uncharacterized isozymes showing distinct amino acid sequences than other organisms. Cyanobacteria are the ancestor of the chloroplast, an endosymbiotic organelle, and most of the enzymes involved in starch metabolism in plants are derived from cyanobacterial homologues23,24. Elucidation of the amylopectin biosynthesis mechanism in cyanobacteria provides clues to understand how amylopectin biosynthesis evolved from glycogen biosynthesis.
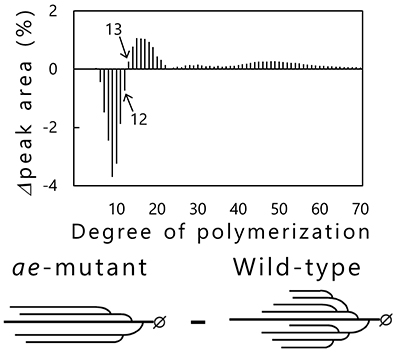
Phenotypic analysis of accumulated starch in the endosperm of rice mutants that are deficient in specific enzymes have been performed to reveal the functions of starch biosynthesis-related enzymes expressed in rice endosperm4,25 (Table 1). BEIIb-deficient mutant rice called amylose-extender (ae)26, for example, accumulates starch exhibiting increased resistant starch content and gelatinization temperature27,28. These properties of the ae mutant are due to longer and fewer branched chains as compared with wild-type rice (Fig. 8). Chain-length distribution patterns of amylopectin in endosperm obtained by subtraction of wild-type rice (japonica variety Kinmaze) from the ae mutant show that the number of short branched chains (DP < 13) is reduced, while that of long branched chains (DP ≥ 13) is increased in the ae mutant, compared with in wild-type rice (Fig. 8). This study proved that the role of BEIIb in starch biosynthesis was cleavage of long branched chains and production of short branched chains26. The role of other rice endosperm isozymes in starch biosynthesis has also been studied by the same method4,25.
A model of amylopectin biosynthesis in rice endosperm has been proposed based on evidence showing that the repeated structure of amorphous lamellae and crystalline lamellae in amylopectin is built by the concerted action of multiple isozymes3,4,12. Recent studies have demonstrated that the amylopectin biosynthesis-related enzymes from terrestrial plants interact with each other to form a protein-protein complex25,29-31. The SSI-SSIIa-BEIIb protein complex has been identified in the rice endosperm (japonica variety Nipponbare)25,30. Although the roles of the protein-protein complexes in starch biosynthesis remain unclear, it is possible that they can supply answers to the following unanswered questions. Functions of starch biosynthesis enzymes have been well characterized both in vivo and in vitro, whereas the mechanism for controlling the structure of amylopectin remains unknown. Several unanswered questions are listed below.
The answers to these questions could allow us to synthesize amylopectins with favorable properties, opening up the possibility for application of novel starch materials such as bioplastics.
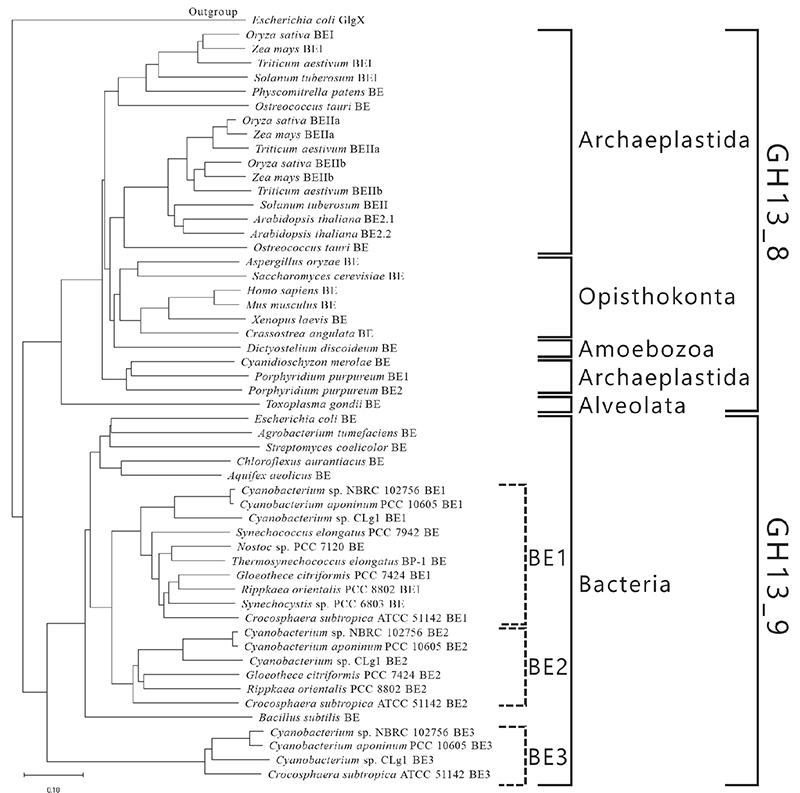
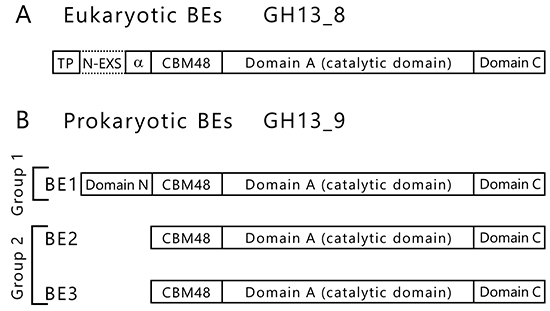
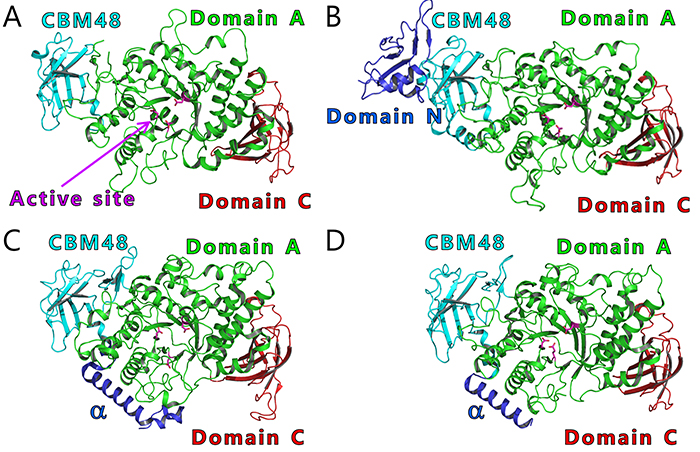
BEs are found in both eukaryotes and prokaryotes and belong to glycoside hydrolase (GH) family 13 (GH13) based on their amino acid similarities32. There are slight differences between the amino acid sequences of eukaryotic and prokaryotic BEs. The eukaryotic and prokaryotic BEs belong to distinct groups, GH13 subfamily 8 (GH13_8) and GH13 subfamily 9 (GH13_9), respectively33, in the phylogenetic tree (with some exceptions)6,7 (Fig. 9). BEs belonging to GH13 basically consist of a carbohydrate-binding module (CBM) family 48 (CBM48), domain A (a catalytic domain), and domain C6,7 (Fig. 10). Eukaryotic BEs (GH13_8) have a long α-helix at their N-termini, whereas prokaryotic BEs (GH13_9) possess (group 1) or lack (group 2) domain N34. Most cyanobacteria have BE1 (a BE belonging to group 1) (Figs. 9 and 10). Amylopectin-producing cyanobacteria possess three isoforms (BE1, BE2, and BE3), and both BE2 (having about 70% amino acid identity to BE1) and BE3 (having about 20% amino acid identity to BE1) belong to group 2 (Figs. 9 and 10).
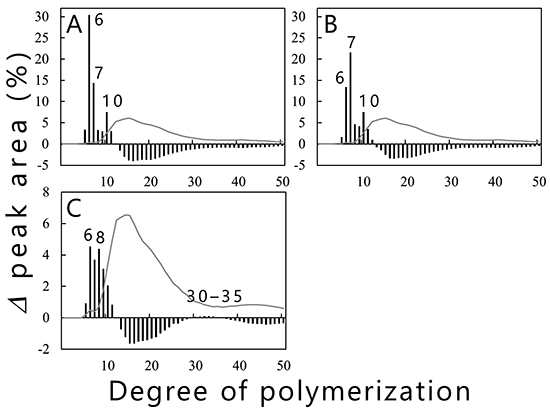
Branched chains in amylopectin influence physicochemical properties such as crystallinity, viscosity, retrogradation rate, and taste. The authors’ studies on cyanobacterial BEs that control the length of branched chains produced by BEs will be described below. Purified recombinant isoforms (cceBE1, cceBE2, and cceBE3) from Crocosphaera subtropica ATCC 51142, an amylopectin-producing cyanobacterium, were prepared and the number and length of the branched chains (chain-length distribution) produced by the three isoforms were analyzed21. The cceBE1 (Fig. 11A) and cceBE2 (Fig. 11B) cleaved branched chains with DP ≥ 12 to produce primarily short branched chains of DP6 and 7. On the other hand, cceBE3 produced branched chains of no preferred length (Fig. 11C), including short branched chains (DP5–11) and a very small amount of long branched chains (DP30–35). The patterns of chain-length distribution produced by BEs from various organisms were shown to be similar to those produced by any of the three isoforms of rice (OsBEI, OsBEIIa, and OsBEIIb)12, and the BEs from various organisms have been classified accordingly into three types, BEI-, BEIIa-, or BEIIb-type35,36. OsBEI, OsBEIIa, and OsBEIIb catalyze the transglucosylation reaction by using branched chains with no less than DP12 as substrates. OsBEIIa produces mainly short branched chains (DP6 and 7) and a lower amount of longer branched chains (DP8–15). OsBEIIb produces short branched chains of DP6 and 7 with very little production of longer branched chains (DP8–15). OsBEI produces short branched chains with DP6–12 as well as long branched chains of DP27–3812. Cyanobacterial BE1 and BE2 can be classified into the BEIIb-type as the patterns of chain-length distribution produced by them are almost identical to that produced by OsBEIIb. The patterns of chain-length distribution produced by cyanobacterial BE3 and OsBEI are similar to each other, and thus the BE3 is grouped with the BEI-type20,21. Prokaryotic and eukaryotic BEs have closely correlated enzymatic properties, although they have been differentiated independently.
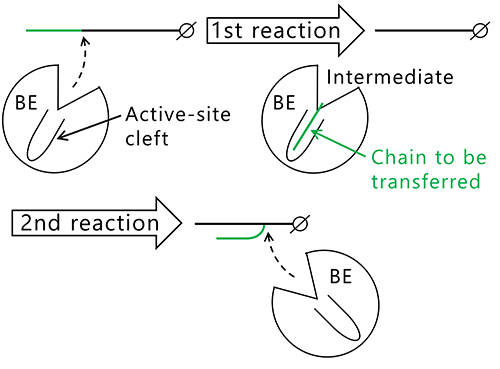
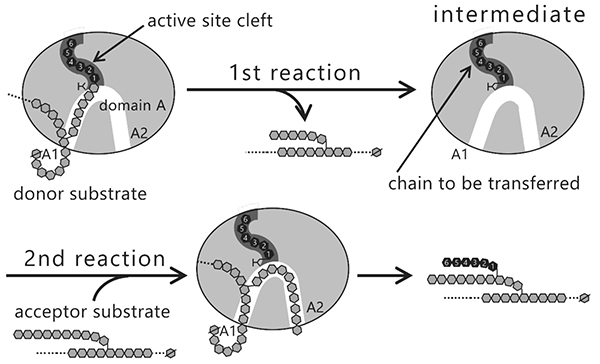
The question that arises here is why the length of the branched chains varies between the isozymes producing them. Reactions catalyzed by BEs are composed of two steps, glycosylation (Fig. 12, 1st reaction) and deglycosylation (Fig. 12, 2nd reaction) reactions7,22,37,38. In the 1st reaction, the α-1,4-linkage in the glucan chain is cleaved and an intermediate is formed where the chain to be transferred is bound to an active site cleft of the BE. In the 2nd reaction, the BE transfers the chain in the intermediate to an acceptor glucan chain via an α-1,6-linkage to form a new branch point. The reaction mechanism employing this type of intermediate is found in amylases and other carbohydrate-active enzymes32,39. Based on the reaction mechanism, it is reasonable to speculate that length of the chain to be transferred is controlled by the shape of active site cleft.
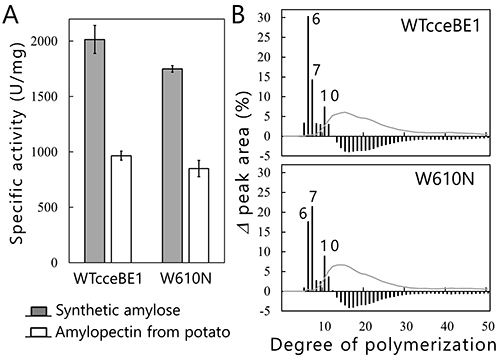
We will test this hypothesis by comparing the crystal structures of BEs. The crystal structures of BEs are available for Escherichia coli40-42 (Fig. 13A), Mycobacterium tuberculosis37 (Fig. 13B), Oryza sativa43,44 (OsBEI) (Fig. 13C), and Homo sapiens45 (Fig. 13D). However, sugar-binding structures of BEs whose active site cleft is occupied by the sugars have never been reported. The authors have overcome this problem by solving the crystal structures of cceBE1 in complex with maltooligosaccharides22. The chain of DP6 to be transferred was found to bind exactly to the active site cleft (Fig. 14A). Next, amino acid residues forming the active site cleft around the non-reducing end of the chain to be transferred (Fig. 14A black circle) were replaced by other amino acids and characterized to investigate the effect of the cleft on catalytic properties of the enzyme. A point mutant, W610N, displaying almost the same activity as wild-type cceBE1 (WTcceBE1) was created22 (Fig. 15A). While WTcceBE1 primarily produces branched chains with DP6 (Figs. 11A and 15B), the major product of W610N was changed to branched chains with DP7 by decreasing the proportion of DP6 (Fig. 15B). It is conceivable that sufficient space for binding of the chains with DP7 were generated at the non-reducing end of the active site cleft of W610N (Fig. 14A, black circle). The crystal structure of W610N in the presence of glucan chains with DP7 (G7) was determined and the active site cleft of the structure was perfectly occupied by the G722 (Fig. 14B). These results strongly suggest that structure of the active site cleft controls the length of chains to be transferred.
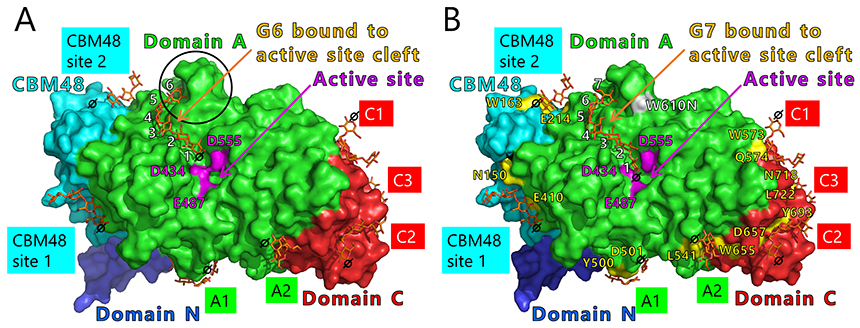
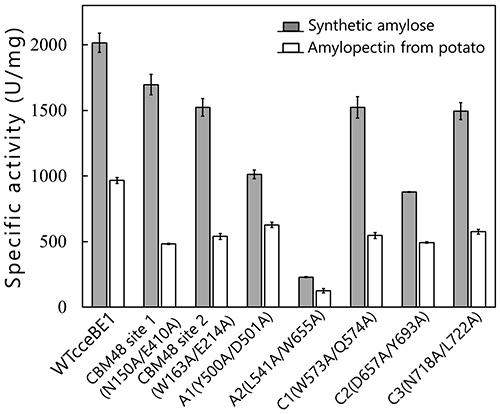
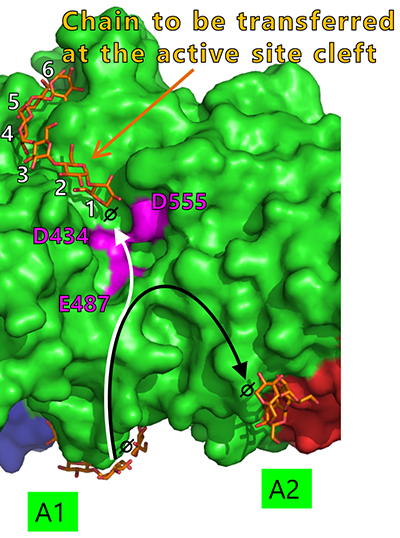
Seven maltooligosaccharide molecules were bound at the surface besides the active site of cceBE1 structures (Fig. 14). Two of the molecules in CBM48 were bound to CBM48 site 1 and CBM48 site 2, respectively46. The others were bound to surface binding sites (SBSs)47,48 and named A1, A2, C1, C2, and C346 (Fig. 14). Seven mutants (in which amino acid residues interacting with bound oligosaccharides [Fig. 14B, yellow] were replaced by alanine) were constructed and characterized. The specific activity of two mutants, A1(Y500A/D501A) and A2(L541A/W655A), was much lower than that of WTcceBE122,46 (Fig. 16), indicating that the A1 and A2 are important for catalysis of BE. The sugars bound at A1, A2, and the active site cleft can be consistently connected from A1 to the active site cleft (Fig. 17, white arrow) and from A1 to A2 (Fig. 17, black arrow). Donor and acceptor substrates are predicted to bind along the pathway specified by the white and black arrows in Fig. 17, respectively. Based on the experimental evidence described above, we propose a BE reaction model22. First, donor substrate binds to the enzyme and extends from A1 to the active site cleft to undergo the 1st reaction (Fig. 18, upper panel). In the 1st reaction, an α-1,4-linkage of the donor substrate is cleaved and the chain to be transferred remains at the active site cleft to form an intermediate that triggers the release of the rest of the substrate from the enzyme (Fig. 18, upper panel). Next, acceptor substrate binds from A1 to A2 giving rise to the 2nd reaction (transfer of the chain to be transferred to the acceptor substrate), which attaches the new chain at a branch point via α-1,6-linkage (Fig. 18, lower panel). The branched structure of polysaccharides produced by BEs can be reasonably explained by this reaction model. The authors proposed this model based on their results of structural analyses22, and are the first to propose such a model.
In this article, the mechanism controlling the length of branched chains produced by BEs was described. Starch can be utilized not only in food production but also in the manufacture of alternative materials such as plastics. Utilization of starch may contribute to the solution of both food shortage problems and global warming issues. Elucidation of the mechanism of starch biosynthesis and control of the structure and properties of starch will enable the practical use of starch, but unanswered questions (see ”6. Effects of biosynthesis-related enzymes on the structure and properties of starch”) remain an obstacle. Further studies are required to provide the answers.
This manuscript (in English and in Japanese) is a revised version of a paper that was orally presented at the 14th Future Forum on Polysaccharides 2020. The work described in this article was supported by grants from the Shimadzu Science Foundation, Japan Foundation for Applied Enzymology, and JSPS KAKENHI (grant numbers 25440193, 15K18685, 16K07467, and 18K06135). We are also grateful to students and researchers for their help in conducting the work.