
Mikiyasu Sakanaka
Program-Specific Associate Professor, Graduate School of Biostudies, Kyoto University, Japan
Bachelor of Engineering at the Department of Bioscience and Biotechnology, College of Science and Engineering, Ritsumeikan University, Japan in 2010. Ph.D. at Graduate School of Agriculture, Hokkaido University, Japan in 2015. Postdoctoral Research Fellow at the Faculty of Bioresources and Environmental Sciences, Ishikawa Prefectural University, Japan in 2015. Specially Appointed Assistant Professor at the Faculty of Bioresources and Environmental Sciences, Ishikawa Prefectural University, Japan in 2017. JSPS Overseas Research Fellow at the National Food Institute, Technical University of Denmark, Denmark in 2018. Program-Specific Associate Professor at the Graduate School of Biostudies, Kyoto University, Japan in 2020. He studies how gut bacteria, including bifidobacteria, have co-evolved with their hosts.
Human milk oligosaccharides (HMOs), which are abundant in human breastmilk, have a prebiotic effect that selectively promotes the growth of bifidobacteria. As a result, breastfed infants often have a bifidobacteria-rich gut microbiota, with bifidobacteria accounting for over 50 % of the total bacterial community. Given this, the application of HMOs to infant formula has begun to make progress in recent years. 2ʹ-Fucosyllactose (2ʹ-FL), a representative species of HMOs decorated with fucose (fucosylated HMOs; FHMOs), was the first HMO species approved for use in infant formula and has attracted attention worldwide. In this chapter, I will describe the diverse FHMO utilization strategies found in bifidobacteria, focusing on their glycosidases and transporters, and discuss the significance of FHMOs applied to fortify infant formula in terms of promoting the formation of a bifidobacteria-rich microbiota.
HMOs are mainly classified into FHMOs, HMOs decorated with sialic acid (sialylated HMOs), and undecorated HMOs1. Among them, FHMOs (Fig. 1) account for ≈70 % (≈7 g/L) of the total HMOs by weight2 and therefore can significantly affect the composition of the infant gut microbiota and encourage the formation of a bifidobacteria-rich microbiota. Interestingly, the abundance and composition of FHMOs vary greatly among individuals, mainly due to the enzymatic activity of the fucosyltransferase-2 (FUT2) that enables α1,2-fucosylation (secretor status)2. Most human mothers (65–98 %) are FUT2 positive donors (secretors) although the proportion can vary across geography2. Secretors can biosynthesize α1,2-FHMOs in the mammary gland, resulting in 2′-FL being the most abundant FHMO species in breastmilk. By contrast, non-secretors harbor both allelic mutations of FUT2 and the corresponding enzyme is inactivated. Therefore, α1,2-FHMOs, including 2′-FL, are absent or limited in quantity in non-secretor breastmilk (Fig. 1), and FHMO species account for only ≈50 % of total HMOs (≈4 g/L)2. Instead, non-secretor breastmilk generally contains an abundant amount of lacto-N-fucopentaose II (LNFP II, an α1,4-FHMO) and/or 3-fucosyllactose (3-FL, an α1,3-FHMO) to compensate lack of α1,2-FHMOs2,3.

Infant-gut associated bifidobacteria are represented by Bifidobacterium longum subsp. longum (B. longum), B. longum subsp. infantis (B. infantis), Bifidobacterium breve, Bifidobacterium bifidum, Bifidobacterium pseudocatenulatum, and Bifidobacterium catenulatum subsp. kashiwanohense (B. kashiwanohense)4. As described in a previous chapter of this series5 (Glycoforum, 24A16), infant-gut associated bifidobacteria are widely known as efficient HMO utilizers, but their FHMO utilization abilities differ greatly between species/strains. Specifically, many strains of B. infantis, B. bifidum, and B. kashiwanohense are capable of efficiently assimilating FHMOs, whereas the abilities of B. longum, B. breve, and B. pseudocatenulatum to assimilate FHMOs vary among strains6. Genomic analyses have also revealed that the gene sets required for FHMO utilization are distributed in almost all strains of B. infantis, B. bifidum, and B. kashiwanohense, but in less than 10 % of strains of B. longum, B. breve, and B. pseudocatenulatum4,7.
Nevertheless, FHMO intake by infants selectively promotes the in vivo proliferation of FHMO-utilizing Bifidobacterium strains, leading to the formation of a bifidobacteria-rich microbiota, although the prebiotic effect of FHMOs can vary among individuals8. Interestingly, breastfeeding by secretor mothers increases the population of bifidobacteria more rapidly and abundantly in the infant gut than breastfeeding by non-secretor mothers9. Furthermore, bifidobacterial strains isolated from the feces of infants breastfed by secretor mothers had relatively high abilities to utilize 2′-FL. These findings indicate that α1,2-FHMOs including 2′-FL promote the selective proliferation of Bifidobacterium strains in infant guts. Berger et al10. then examined the prebiotic effect of 2′-FL (and lacto-N-neo-tetraose [LNnT; a non-fucosylated HMO species]) applied to fortify infant formula. Their results showed that bifidobacteria were more abundant in the guts of infants fed 2ʹ-FL/LNnT-supplemented formula than those fed a control formula. This is the first demonstration showing the prebiotic effect of HMO species in terms of infant gut microbiota. Although the variety of HMO species applied to infant formula is still limited, future application of additional HMO species will contribute to the development of an infant formula closer to the composition of breastmilk.
The previous sections have described how FHMOs can promote the in vivo proliferation of bifidobacteria in infant guts. To further maximize the prebiotic effect of various FHMO species that may be applied to infant formula in the future, a molecular understanding of FHMO utilization strategies in bifidobacteria is required. As described in a previous chapter of this series5, B. bifidum has been reported to have two distinct extracellular fucosidases (1,2-α-ʟ-fucosidase and 1,3/4-α-ʟ-fucosidase) and other several glycosidases to extracellularly degrade almost all types of FHMOs11-13. The extracellular glycosidase activity is a common feature among B. bifidum strains, as homologs of the respective genes are found in almost all strains of this species4,14. Interestingly, in vitro studies have demonstrated that fucose and other mono- and disaccharides (FHMO degradation products) liberated extracellularly by B. bifidum are not only partially utilized by B. bifidum itself but also cross-fed to other infant-gut associated Bifidobacterium species that are unable to utilize FHMOs14. This suggests that B. bifidum-mediated cross-feeding of FHMO degradation products promotes the formation of a bifidobacteria-rich microbiota, indicating that B. bifidum plays a crucial role in the infant gut ecosystem. However, recent metagenomic analyses have shown that the prevalence of B. bifidum in breastfed infant guts is not as high as that of other infant-gut associated Bifidobacterium species8,9,15-18. Although the reason is unclear, it may be partly because B. bifidum is more negatively affected by antibiotics administered to infants than other Bifidobacterium species17.
In contrast to B. bifidum, other FHMO-utilizing Bifidobacterium species and strains have strategies to degrade FHMOs intracellularly4. They not only are equipped with intracellular fucosidases (1,2-α-ʟ-fucosidase and 1,3/4-α-ʟ-fucosidase) and other several glycosidases, but also possess a transporter(s) to import FHMOs (FL transporter)4. The FL transporter gene homologs are distributed in only a few strains of Bifidobacterium19 and are classified into four clusters based on phylogenetic analysis (clusters 1-IV, 2-I, 2-II, and 2-III; the amino acid sequences differ by up to 40 % among the clusters)19,20. Recent studies have shown that the substrate specificities of these FL transporters differ markedly among the clusters, indicating the diversification of FHMO utilization strategies in bifidobacteria8,19-23. As summarized in Fig. 2, the FL transporters belonging to clusters 1-IV can import only 2′-FL and 3-FL, whereas clusters 2-I and 2-II can import not only 2′-FL and 3-FL, but also lactodifucotetraose (LDFT) and lacto-N-fucopentaose I (LNFP I). Furthermore, cluster 2-III can take up the widest range of FHMO species, 2′-FL, 3-FL, LDFT, LNFP I, LNFP II, and lacto-N-difucohexaose I/II (LNDFH I/II) (Fig. 2), showing better adaptation to various FHMOs from both secretor and non-secretor mothers than other FL transporter clusters (clusters 1-IV, 2-I, and 2-II) (Figs. 1, 2). It should be noted that the types of FL transporter clusters differ remarkably among species and strains (Fig. 2)20, which is consistent with the above-mentioned differences in FHMO utilization abilities among strains.
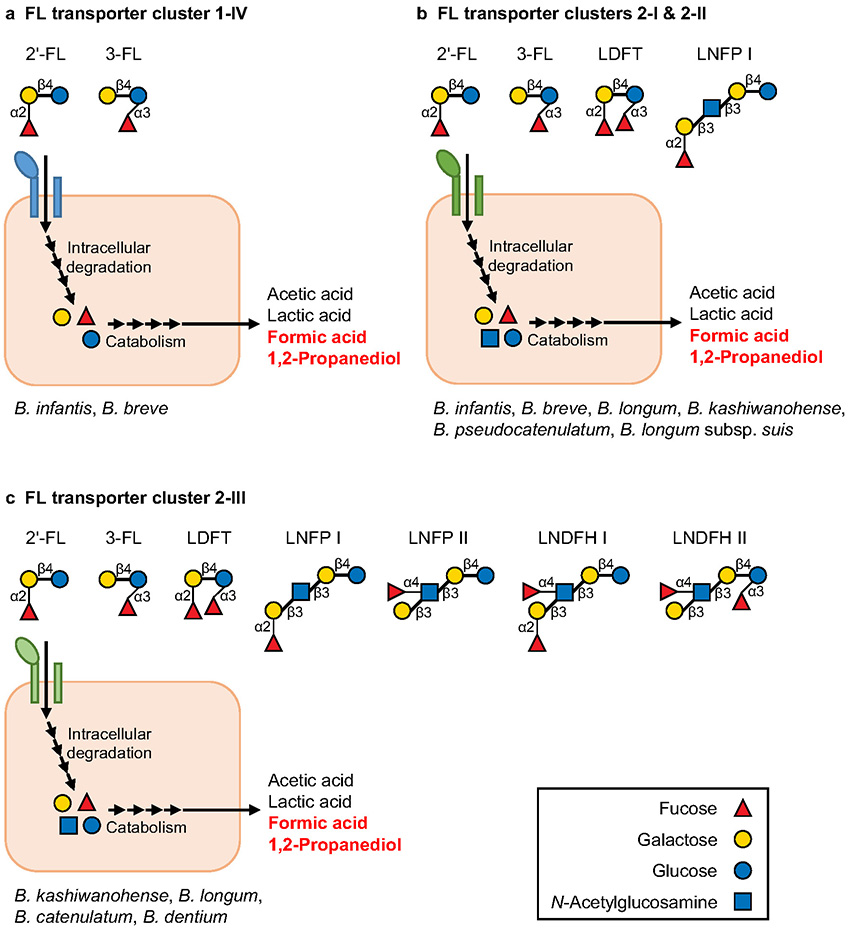
The above-mentioned studies on FHMO utilization (intracellular or extracellular FHMO degradation) have mainly been conducted in vitro, but recent in vivo analyses using mouse models and infant feces have also demonstrated that FHMO utilization genes are closely associated with the formation of a bifidobacteria-rich microbiota. For example, in a 2ʹ-FL-fed mouse model, an FL transporter positive strain (B. pseudocatenulatum MP80) was shown to proliferate more efficiently than an FL transporter negative strain of the same species in the mouse gut30. Additionally, while the effects of secretor status are unclear, FL transporter genes (especially, clusters 2-I, 2-II, and 2-III; Fig. 2) were found to be more abundant in the feces of breastfed infants than in adults, and the abundance of these genes was shown to be positively correlated with the abundance of bifidobacteria in the infant feces and negatively correlated with the concentration of FHMOs remaining in the feces19. By contrast, the FL transporter genes were scarcely detected in the feces of infants fed formula lacking HMOs, and as reflected in this observation, the abundance of bifidobacteria was relatively low. These results suggest that the FL transporter-mediated utilization of FHMOs considerably contributes to the in vivo proliferation of bifidobacteria in infant guts. However, the FL transporter (intracellular FHMO degradation) is not the sole determinant for bifidobacteria-rich microbiota formation, i.e., the microbiota formation should be partly attributed to B. bifidum-mediated cross-feeding of FHMO degradation products (extracellular FHMO degradation). In fact, the gene set containing extracellular fucosidase (B. bifidum) and fucose transporter FucP (other Bifidobacterium species) are detected in the feces of some breastfed infants with an FL transporter negative bifidobacteria-rich microbiota16. These results strongly suggest that FHMO degradation products (mono- and disaccharides) liberated by B. bifidum are cross-fed to other FL transporter negative Bifidobacterium species in some breastfed infants. However, as mentioned earlier, considering that B. bifidum prevalence is comparatively low in breastfed infant guts and most other Bifidobacterium strains are FL transporter negative, the prebiotic effect of FHMOs on the in vivo proliferation of bifidobacteria can vary among infants8,10. To maximize the future application of various FHMOs for formula fortification, prior examination of the fecal abundance of FHMO utilization genes (e.g., FL transporter or extracellular fucosidase genes) and/or combined intake of FL transporter positive Bifidobacterium strains and FHMOs may be required.
In addition to the mechanisms of bifidobacteria-rich microbiota formation (FHMO utilization), the molecular mechanisms by which bifidobacteria exert beneficial effects on infant health is attracting worldwide attention. It has long been known that sugar (HMO) catabolism by bifidobacteria leads to the production of acetic acid and lactic acid as major short-chain fatty acids, which have beneficial effects such as preventing pathogen infection by lowering the pH of the infant gut environment31. Interestingly, several in vitro studies have reported the catabolism of fucose from FHMOs to contribute to a unique metabolite profile, i.e., high production of formic acid and 1,2-propanediol in addition to acetic acid and lactic acid22,24-26 (Fig. 2). In vivo concentration of formic acid has also been confirmed to be considerably high when the FHMO utilization genes are abundantly present in infant feces16. The 1,2-propanediol produced by bifidobacteria has been shown to not only be cross-fed to gut bacteria such as Eubacterium hallii and lactobacilli27,32 but also promote host immune responses33. Although the ecological and physiological significance of formic acid and 1,2-propanediol in the infant guts remains unclear, further research may help clarify why a limited number of Bifidobacterium strains have adapted to metabolize FHMOs.
During the last several years, fortification of infant formula has made rapid progress in terms of supplementation with HMO species. Although HMOs applied are currently limited to several species, the variation is expected to more increase in the near future. Thus, it is important to elucidate the molecular mechanisms by which gut bacteria utilize various HMO species, especially FHMOs. As described in this chapter, various studies over the past 15 years have revealed the detailed utilization strategies of FHMOs by bifidobacteria. Recent studies have also reported the utilization of FHMOs in some other gut bacterial species such as Bacteroides spp., Akkermansia muciniphila, and Clostridiales34-36. These findings suggest that the utilization of FHMOs in the infant guts is much more complex than initially expected. Thus, further research clarifying the function and role of each enzyme and transporter in various gut bacteria, including bifidobacteria, will be required to grasp the whole picture of FHMO utilization strategies in infant gut-associated bacteria.
I am grateful to Dr. Miriam N. Ojima for carefully reading the manuscript.