
Toshihiko Katoh
Associate Professor, Graduate School of Biostudies, Kyoto University
Dr. Katoh graduated from the Department of Biofunctional Science, Faculty of Agriculture, Kyoto University in 2001. He obtained a doctoral degree in life science from the Graduate School of Biostudies, Kyoto University under the tutelage of Professor Kenji Yamamoto in 2007. He served as a postdoctoral researcher under Professor Michael Tiemeyer at the University of Georgia Complex Carbohydrate Research Center until 2013. He was appointed as an assistant professor at the Research Institute for Bioresources and Biotechnology, Ishikawa Prefectural University in 2013, and as an assistant professor in the Graduate School of Biostudies, Kyoto University in 2015. He has been in his current position since 2023. His research interests include interactions between human enteric bacteria and host-derived glycoconjugates.

Takane Katayama
Professor, Graduate School of Biostudies, Kyoto University
Takane Katayama is a Professor at Kyoto University. He received his Ph.D in the field of applied molecular microbiology under the supervision of Prof. Hidehiko Kumagai. After spending three years as a postdoc in the same lab, he moved to the lab led by Prof. Kenji Yamamoto and was appointed an assistant professor. In Prof. Yamamoto’s lab, he isolated the genes for 1,2-α-L-fucosidase and endo-α-N-acetylgalactosamindase from bifidobacteria, both of which are enzymes acting on human-derived glycans. These findings prompted him to consider host glycans-mediated symbiosis between gut microbes and humans. In the last decade, he has focused on functional analysis of bifidobacterial genes and enzymes involved in degradation of human milk oligosaccharides. His research has significantly contributed to revealing how bifidobacteria-rich microbiota is formed in the gut of breast-fed infants.
Abbreviations
CBM, carbohydrate-binding module; Fuc, fucose; Gal, galactose; GalN, galactosamine; GalNAc, N-acetylgalactosamine; GH, glycoside hydrolase; GlcNAc, N-acetylglucosamine; GlcNAc-6S, 6-O-sulfated N-acetylglucosamine; LacNAc, N-acetyllactosamine; Neu5Ac, N-acetylneuraminic acid.
Certain gut bacteria produce mucin-degrading enzymes as part of their strategy to adapt themselves to the human intestine. Recent advances in the analysis of these enzymes have identified a wide variety of enzymatic repertoires corresponding to the complex structures of mucins. In addition, information about such enzymes has been compiled in various databases, making it easier to provide a comprehensive understanding of mucin decomposition and related metabolic pathways in the human intestine. This paper discusses mucin decomposition pathways derived from the characteristics of mucin-decomposition-associated enzymes that have so far been identified in gut bacteria.
Intestinal mucosal tissue is the anatomical site not only for digestion and absorption, but also for symbiosis with enteric bacteria, especially in the large intestine. Mucins secreted from the gastrointestinal epithelium in large amounts play roles in forming a mucus layer to protect the tissue, capturing bacteria, viruses, etc., and eliminating them from the body1. While some pathogenic bacteria decompose and erode this mucin mucus layer and invade the host’s body, it has been found that a range of nonpathogenic indigenous bacteria and probiotics (often termed “good bacteria”) have mucin-degrading activity and proliferate in the intestine by assimilating and utilizing mucins2. Furthermore, as mucin decomposition products, free saccharides are shared and metabolized by the bacterial flora into useful moieties such as short-chain fatty acids; mucin assimilation is thus also considered to be important for the host’s health and the maintenance of a balanced bacterial flora (eubiosis). Therefore, in discussing the influence of mucin decomposition on the host, it is necessary to have two viewpoints, i.e., pathology and symbiosis. To gain a comprehensive understanding, however, it is important to understand the mucin decomposition pathways of various bacteria in relation to mucin decomposition activity. This paper describes enzymatic repertoires possessed by major mucin-assimilating enteric bacteria and discusses their likely mucin decomposition pathways.
There are two major types of mucin: secretory gel-forming mucins and membrane-associated mucins with a transmembrane domain. Both types have a mucin domain rich in proline, threonine, and serine. Mucins are characterized by the dense addition of O-linked glycans (mucin-type glycans, O-glycans) with α-linked GalNAc bound to the hydroxyl group of the threonine or serine residue in the mucin domain. Found in the intestine of healthy humans, O-glycans have core 1–4 structures and undergo elongation by type-1/2 LacNAc structure and modifications with fucose, sialic acid, sulfate groups, and acetyl groups to assume extremely heterogenous diverse structures, thus presenting a wide variety of terminal carbohydrate antigens (Figure 1). For details of O-glycan biosynthesis, refer to the literature3.
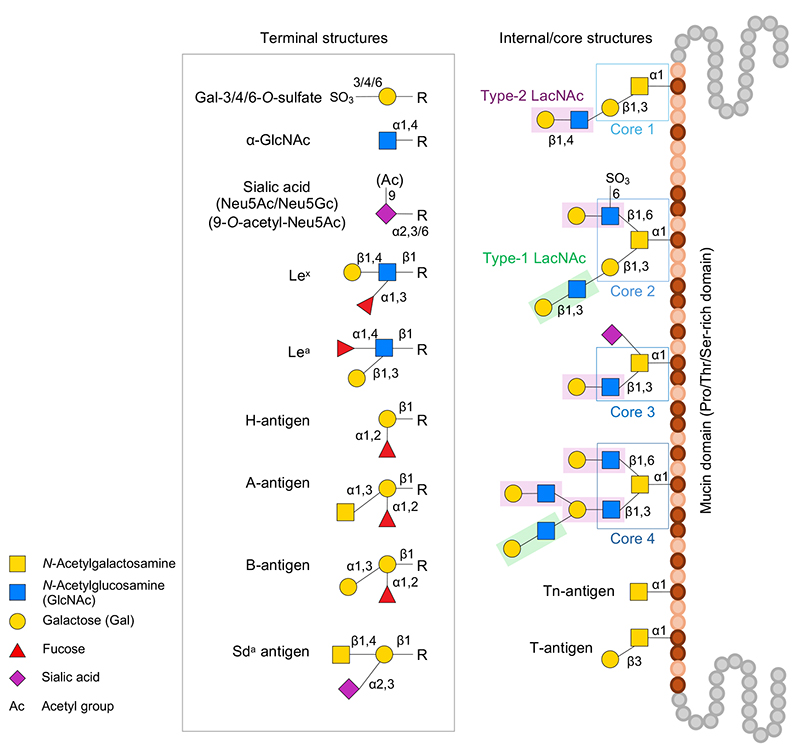
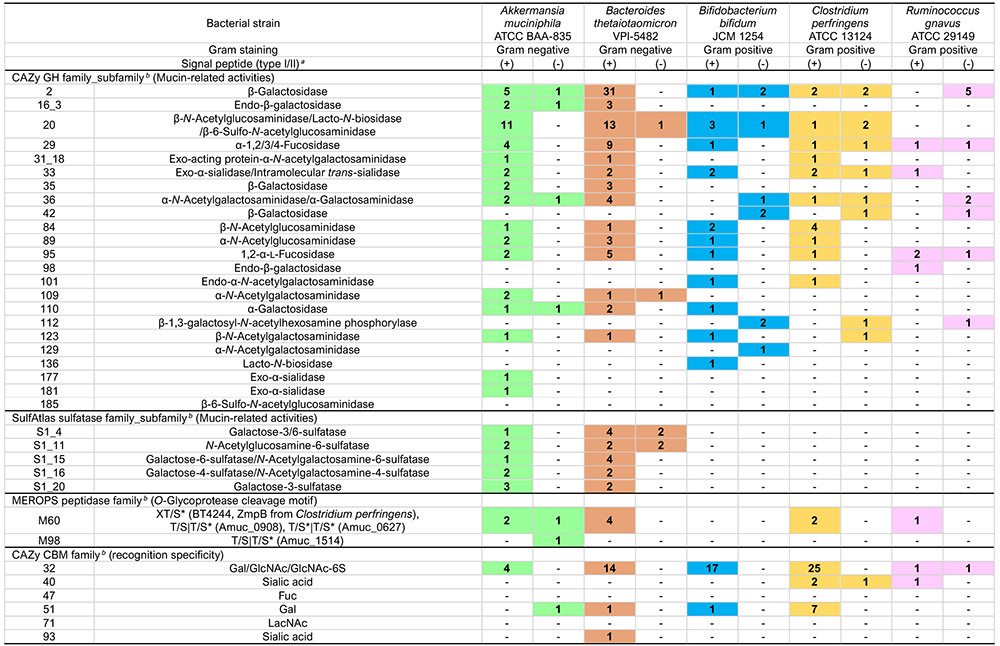
Glycoside hydrolases (GH, EC3.2.1.-) are classified into 187 families in the CAZy database (http://www.cazy.org/)4 (as of December 2023). At least 23 GH families or subfamilies have been experimentally shown to be involved in the decomposition of mucin oligosaccharides or are considered to be possibly involved (Table 1).
The following GH enzymes are involved in the decomposition of carbohydrate terminal modifiers. GH95 1,2-α-ʟ-fucosidase is involved in H-antigen (Fucα1-2Galβ1-R), and GH29 1,3/4-α-ʟ-fucosidase is involved in the release of the fucose contained in Lewis antigens5-8. Involved in the release of sialic acid are GH33 and GH181 exo-α-sialidase8-10. GH177 exo-α-sialidase was first identified in the periodontal disease-causing bacterium Tannerella forsythensis11 and is also found inAkkermansia and other bacteria; however, it remains unknown whether this enzyme is active on mucins. The GH33 enzyme from Ruminococcus gnavus has intramolecular trans-sialidase activity, acting specifically on α-2,3-bound Neu5Ac to produce and release 2,7-anhydro-Neu5Ac12,13. The α-N-acetylglucosaminidase of GH89 acts on the α-linked terminal GlcNAc residue14,15. GH36/GH109 α-N-acetylgalactosaminidase and GH110 α-galactosidase act on blood group A or B antigens (GalNAcα1-3(Fucα1-2)Galβ1-R and Galα1-3(Fucα1-2)Galβ1-R), respectively to convert the antigens to H-antigens16,17. There is another known pathway in which GalNAc deacetylase acts on the GalNAc residue of blood group A antigen to convert the residue to GalN, after which GH36 α-galactosaminidase acts to decompose the GalN18. In addition, GH98 endo-β-galactosidase releases blood group A/B antigen trisaccharides19.
The decomposition of type-2 LacNAc (Galβ1-4GlcNAcβ1-R) is mediated by GH2 β-galactosidase and GH20/GH84 β-N-acetylglucosaminidase20,21. While the decomposition of type-1 LacNAc (Galβ1-3GlcNAcβ1-R) is considered to be mediated by GH35 β-galactosidase22, it also occurs with GH20/GH136 lacto-N-biosidase. In the latter case, the disaccharide lacto-N-biose I (LNB) is released23–25. Furthermore, GH42 β-galactosidase may catalyze the decomposition of the type-1/2 LacNAc structure in cells26,27. GH16_3 endo-β-galactosidase acts on the β-Gal bond in O-glycan to release oligosaccharides, and this reaction reportedly requires the removal of the sialic acid from the carbohydrate terminal in advance28.
The core 1 structure (T-antigen, Galβ1-3GalNAcα1-Ser/Thr), like type-1 LacNAc, is thought to be converted to Tn-antigen (GalNAcα1-Ser/Thr) by the action of GH35 β-galactosidase22. In addition, GH101 endo-α-N-acetylgalactosaminidase acts on the T-antigen to release the disaccharide Galβ1-3GalNAc (galacto-N-biose, GNB)29,30. The core 2 structure (Galβ1-3(GlcNAcβ1-6)GalNAcα1-Ser/Thr) is converted to the core 1 structure by the action of GH84 β-N-acetylglucosaminidase21. Conversely, core 2 structure (Galβ1-3(6-sulfo-GlcNAcβ1-6)GalNAcα1-Ser/Thr), which contains 6-sulfated GlcNAc (GlcNAc-6S), is converted to core 1 by the action of GH20 6-sulfo-β-N-acetylglucosaminidase (sulfoglycosidase)31. GH185 6-sulfo-β-N-acetylglucosaminidase may also mediate the decomposition of GlcNAc-6S residues in mucin carbohydrates32. The core 3 structure (GlcNAcβ1-3GalNAcα1-Ser/Thr) is converted to Tn-antigen by the action of GH20 β-N-acetylglucosaminidase21. The Tn-antigen releases GalNAc by the action of GH31_18/GH129 α-N-acetylgalactosaminidase33,34.
While the members of the GH family are classified according to amino acid sequence homology, members of the same family often include more than one activity with different specificities, and can also exhibit unknown substrate specificity. It should be noted, therefore, that the presence of such GH in the genome does not always imply that it acts on mucin.
SulfAtlas (https://sulfatlas.sb-roscoff.fr/)35 is available as a database for sulfatases (sulfohydrolase, EC3.1.6.-). The database classifies sulfatases into four families, S1 to S4, and many subfamilies. S1, specifically, comprises 110 subfamilies (each represented by S1_X). Members have carbohydrate sulfatase activity, acting on sulfated saccharides that belong to the S1 family. The active center of sulfatase is the Cα-formylglycine residue that results from translationally coupled modification of Cys or Ser residue36, and this reaction is catalyzed by anaerobic sulfatase-maturating enzyme (anSME) in anaerobic bacteria.
Sulfate group modifications of mucins are found at the 3-, 4-, and 6-positions of Gal, the 6-position of GlcNAc, the 8-position of Neu5Ac37, and elsewhere. Martens et al. provide a great deal of information about the enterobacterial sulfatase from Bacteroides thetaiotaomicron38,39. B. thetaiotaomicron carries a great many (28) S1 sulfatase genes that are present in a cluster of genes related to the utilization of polysaccharides such as mucins and glycosaminoglycans (polysaccharide utilization loci, PUL). Mucin-related PULs that cluster with SusC/SusD (a system in which bacteria incorporate polysaccharide metabolites into bacteria), GH, etc. include S1_4, S1_11, S1_15, S1_16, and S1_2040.
S1_4, S1_15, S1_16, and S1_20 act on sulfated Gal, of which S1_4 and S1_20 exhibit galactose-3-sulfatase activity, acting on 3-sulfated Gal; and S1_15 exhibits galactose-6-sulfatase and N-acetylgalactosamine-6-sulfatase activity, acting on 6-sulfated Gal or 6-sulfated GalNAc, as shown in Table 1. S1_16 exhibits galactose-4-sulfatase and N-acetylgalactosamine-4-sulfatase activities against 4-sulfated Gal (GalNAc). S1_11 exhibits specificity for 6-sulfated GlcNAc (GlcNAc-6S). In addition, the S1_53 enzyme from Bifidobacterium breve is reportedly involved in the metabolism of GlcNAc-6S41. As far as the author knows, no sulfatase has been identified to act on sulfated sialic acids.
The term O-glycoprotease refers to a protease (peptidase) that acts on proteins with O-glycans. O-glycoproteases, such as OgpA and CpaA, exhibit a broad range of specificity for glycoproteins with O-glycans. The present study focuses on O-glycoproteases that act specifically on glycoproteins with mucin domain(s), also known as mucinases due to their recognition of mucin domain structures and possession of the carbohydrate-binding modules (CBMs) described below42. Not only pathogenic but also indigenous enteric bacteria secrete mucinase to decompose mucin polypeptides. These enzymes are for the most part extracellular zinc-requiring metalloproteases classified as certain peptidase families43 (M60, M66, M98, etc.) that constitute the M60-like family (Pfam 13402)44 in the MEROPS database of peptidases and their inhibitors. The members of the M60-like family share the HEXXH motif in their domains. The two histidine residues in the motif are involved in the binding of zinc ions, and the glutamate residue is a catalytic residue. Found outside the M60-like family is the O-glycoprotease activity in serine protease.
Although the substrate pocket of O-glycoprotease recognizes different carbohydrate structures depending on the enzyme, carbohydrate trimming by GH is thought to promote the decomposition of O-glycoprotease because many such enzymes recognize relatively short-trimmed O-glycans of (sialyl)T- and (sialyl)Tn-antigens45. Some members of the M60 family are known to be associated with pathogenicity46, and this is attributable to the fact that they directly decompose the mucin polypeptide chain that constitutes the mucin layer backbone, to facilitate the entry of pathogenic bacteria into epithelial cells.
Recently, there has been rapid development of practical applications of O-glycoproteases, focusing considerable attention on their use as analytical tools for extensive structural analyses of mucin-domain-containing glycoproteins47 and mucin staining45.
Carbohydrate-binding modules (CBMs), which is “defined as a contiguous amino acid sequence within a carbohydrate-active enzyme with a discreet fold carbohydrate-binding activity” (CAZy database), are currently classified into 100 families in the CAZy database. CBMs considered to be involved in the recognition of mucin carbohydrates include CBM32, CBM40, CBM47, CBM51, CBM71, and CBM93. In particular, CBM32 is associated with the recognition of Gal, GalNAc, GlcNAc, and GlcNAc-6S and is found in many GHs and OO-glycoproteases. The binding to mucin carbohydrates by CBMs enables the direct contact of the enzyme to the mucin, resulting in improved mucin decomposition efficiency31.
Other enzymes are known to be associated with mucin decomposition, including sialate-O-acetylesterase, which works in the decomposition of 9-O-acetylated sialic acid, and LNB/GNB phosphorylase (GH112), which phosphorolyzes LNB or GNB48. Sialate-O-acetylesterase is sometimes found as a multi-domain enzyme working together with sialidase9. Because the known LNB/GNB phosphorylase is an intracellular enzyme, no cases of direct action on mucins have been found.
Because the retention pattern for the enzymes described above differs depending on the bacterial species and strain, the mucin decomposition pattern is also non-uniform. In addition, as described above, it is necessary to empirically confirm the same activity and specificity even when employing enzymes of the same family. Furthermore, even when enzymes exhibit the same activity, the decomposition pathway and their influence on other bacteria are likely to vary widely among different situations, such as extracellular action, transportation as an extracellular vesicle cargo protein, and action in the periplasmic space or cytoplasm. Therefore, it is necessary to consider not only the identity of the enzyme, but also the localization of each enzyme, when discussing the mucin decomposition pathway of each bacterium.
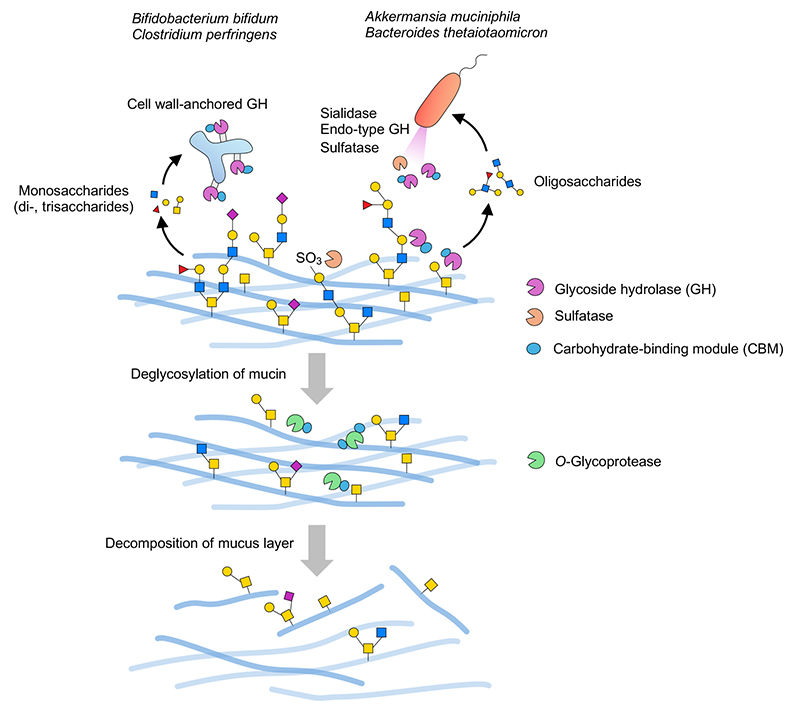
Gram-negative bacteria of the genus Bacteroides and other species are known to have many PULs as described above. In the plant-derived polysaccharide utilization pathway, endo-type secretory enzymes often decompose polysaccharides into oligosaccharides extracellularly, these then being transported to the periplasmic space by the SusC/SusD uptake system. Oligosaccharide-degrading enzymes are present in the periplasmic space, where the oligosaccharides are decomposed to monosaccharides. Finally, the monosaccharides are transported to the cytoplasm and assimilated. This system is considered to apply to mucin carbohydrates as well. According to a review by Luis et al.40, the PUL database (PULDB)4, and other sources, the mucin PUL of B. thetaiotaomicron is not only coded with sulfatases such as S1_20/S1_11, but also with GH2/GH16_3/GH20/GH29/GH95 and others. It is conjectured that secretory GH33 sialidase (BT0455)28,49 or S1_20 galactose-3-sulfatase (BT1636), which is assumed to be a lipoprotein localized in the outer membrane or in outer-membrane-derived-vesicles38, first acts directly on the mucin extracellularly to remove acidic terminal-modifying residues such as sialic acid residues and sulfate groups, respectively, followed by the actions of extracellular endo-type enzymes such as GH16_3 endo-β-galactosidase to produce oligosaccharidesspan class="sld">28. The oligosaccharides are transported to the periplasmic space by SusC/SusD on the outer membrane periplasm, where they are further decomposed by periplasmic localized sulfatases and GHs40. Intake of mucin-derived oligosaccharides in the periplasm appears to be important in the efficient assimilation of these saccharides. Once cropped by GH16_3 endo-β-galactosidase, mucin glycoproteins are likely to serve as a good substrate for O-glycoprotease because they are considered to expose the core structure and short glycan structures around it (Figure 2).
On the other hand, some Gram-positive bacteria with no periplasmic space (e.g. Bifidobacterium and Clostridium) have cell-wall-anchored GHs. These GHs are once biosynthesized as membrane proteins and then immobilized onto the peptidoglycan layer by covalent bonding via a sortase-dependent pathway50. As such, the GHs localized on the cell surfaces sequentially decompose mucin carbohydrates from their terminals to release monosaccharides and short oligosaccharides into the environment (Figure 2), and available saccharides are absorbed into bacterial cells. Because the enzymes are presented on the bacterial cell surface layers, it is thought that the bacterial cells must adhere to the mucin in order to efficiently decompose it. In this regard, it worth noting that many CBMs are present in cell-wall-anchored GH enzymes. These CBMs can bind their substrates, playing a key role in improving the action profile of the catalytic domain31. As giant molecules, mucins are not highly dispersible in liquids. Therefore, capture of the substrate on the mucin by CBM is considered essential, markedly increasing the efficiency of the association between the substrate and the enzyme. CBMs are also found in Bacteroides- and Akkermansia-derived enzymes; however, extracellular enzymes that act directly on mucins often appear to have CBMs. The mechanism of mucin decomposition by GH identified as an extracellular vesicle cargo protein remains to be elucidated by future analyses.
Based on genomic information from mucin-assimilating bacteria, a statistical examination of the association between the identity of each CBM family and their numbers, and the presence or absence of various GH families, revealed the interesting finding that GH16_3 endo-β-galactosidase may have the greatest influence on the retention of CBM31. Lacking GH16_3, Gram-positive bacteria, such as Bifidobacterium bifidum and Clostridium perfringens, for the most part have high CBM retention rates, whereas Gram-negative bacteria with GH16_3 have low CBM retention rates. This relationship is reasonable as oligosaccharides that have been cleaved by an endo-type enzyme show high enzyme association efficiency even in the absence of CBM because they are highly dispersible in liquids. This led to the hypothesis that there are two decomposition modes in the enterobacterial mucin decomposition pathway: CBM-dependent and endo-type-enzyme-dependent. Interestingly, some strains of C. perfringens presumably have an endo-type-enzyme-dependent pathway, with different mucin decomposition modes found in the same species. In addition, Ruminococcus gnavus, a Gram-positive bacterium, may decompose mucin in a mode distinct from the above two pathways as it has a low CBM retention rate and lacks GH16_3. Alternatively, it may exhibit only limited mucin-degrading activity because the number and variety of mucin-degrading enzymes is relatively small.
The mucin carbohydrate decomposition mechanism of Bifidobacterium bifidum, which has been the focus of our investigation, is outlined below. It is known as a probiotic organism and often detected in both the adult and infant gut. It is also known to decompose and metabolize human milk oligosaccharides, which are structurally similar to mucin O-glycans51.
Most steps in the mucin decomposition by B. bifidum occur extracellularly by the action of the aforementioned cell-wall-anchored GH. Fucose is released from H-antigens and Lewis antigens by the action of GH95 1,2-α-L-fucosidase (AfcA)5 and GH29 1,3/4-α-L-fucosidase (AfcB)6, and sialic acid is released by the action of GH33 sialidases (SiaBb19,52, SiaBb210,52,53). SiaBb1 is a multi-domain enzyme with not just a GH33 domain but also an SGNH hydrolase domain9, which exhibits sialate-O-acetylesterase activity. Yet another multi-domain enzyme (SiaBb3) was found to have GH123 β-N-acetylgalactosaminidase and sialidase domains and is considered to mediate the decomposition of Sda antigen (Neu5Acα2,3 (GalNAcβ1,4)Galβ-R) (Ashida et al., unpublished data). GH89 α-N-acetylglucosaminidase AgnB acts to release the α-GlcNAc cap (terminal α1,4-linked GlcNAc moiety, as is often found in gastric mucin etc.)15. While B. bifidum has GH110 α-galactosidase AgaBb54 as an enzyme that acts on blood group B antigens, no GH109 α-N-acetylgalactosaminidase has been identified to act on blood group A antigens, and the bacterium does not carry homologues of GalNAc deacetylase and GH36 α-galactosaminidase. In addition, GH2 β-galactosidase BbgIII may be involved in the decomposition of type-2 LacNAc, and GH20 β-N-acetylglucosaminidase BbhI and GH84 β-N-acetylglucosaminidase BbhIV may be involved in the decomposition of β-GlcNAc residues20,21. GH20/GH136 lacto-N-biosidase acts in the decomposition of Type-1 LacNAc to release the disaccharide LNB23,55,56. B. bifidum has no sulfatase gene for sulfated sugars; however, β-GlcNAc-6S bonds are decomposed by GH20 sulfoglycosidase BbhII to release sulfated GlcNAc31. There is no identified enzyme involved in the decomposition of sulfated Gal. The GH84 β-N-acetylglucosaminidase BbhIV acts on the β1,6-GlcNAc residues contained in the core 2 and core 4 structures to convert these structures to core 1 and core 3, respectively21. GH20 β-N-acetylglucosaminidase BbhI acts on the β1,3-GlcNAc residue in core 3 to convert core 3 to the Tn-antigen21. GH101 endo-α-N-acetylgalactosaminidase is thought to act on the core 1 structure (T-antigen) to release the disaccharide GNB. The released LNB and GNB are transported to the cytoplasm by ABC transporters and phosphorolyzed to Gal-1-phosphate and GalNAc/GlcNAc by GH112 GNB/LNB phosphorylase LnpA1 or LnpA248. Localized in the cytoplasm, GH129 α-N-acetylgalactosaminidase NagBb acts on Ser/Thr or glycopeptides with a Tn-antigen, releasing GalNAc34. However, the mechanism by which glycoamino acids and glycopeptides with a Tn-antigen are produced and transported into the cells remains to be clarified.
As described above, B. bifidum has a GH repertoire corresponding to the diverse glycan structures found in mucin, adapting itself at high levels to survive in the environment of the human intestinal mucin layer. Interestingly, this bacterium has GH16 gene fragments in its genome. However, because the bacterium lacks the active center of GH16, its gene products are not considered to be enzymatically active. This may represent an evolutionary record of how the bacterium selected the CBM-dependent mucin decomposition strategy, rather than the GH16_3 endo-enzyme-dependent strategy, in the history of bacterial evolution that led to adaptation to the mucin layer.
As described above, many enzymes have been reported to involve mucin decomposition, with their information now compiled in various databases. Further as-yet-unknown enzymes will be identified, and new activities and specificities will be discovered. Such findings are expected to facilitate a more profound understanding of biological interactions and lead to the development and establishment of effective solutions for the increase in gastro-intestinal disorders seen around the world.