Tetsuya Okajima
Professor, iGCORE, Graduate School of Medicine, Nagoya University
Graduated from Nagoya University School of Medicine in 1996 and completed the doctoral program at the graduate school of medicine, Nagoya university in 2000, earning a Ph.D. in Medicine. After studying at the Ken Irvine Laboratory at Rutgers University in the United States, he became an assistant professor at the graduate school of bioagricultural sciences, Nagoya University in 2005 (Matsuda Lab). In 2008, he became an associate professor at the center for neurological diseases and cancer, Nagoya university graduate school of medicine (Furukawa Lab), and in 2015, he became a full professor at the same institution. Since 2018, he has concurrently served as a professor at iGcore. In 2010, he received the Japan Biochemical Society Encouragement Award. He continues to explore new functions of glycans through functional analysis of atypical O-glycan modifications of NOTCH receptors. He is involved in promoting collaborative research with researchers from different fields through cohort studies at HGA.
Standardized analytical methods have been established for genome and proteome analyses, enabling large-scale studies such as whole-genome analyses and comprehensive protein profiling, which have revealed associations with major diseases1-5. Meanwhile, the development of analytical platforms for glycan profiling by the Human Glycome Atlas Project has made it possible to conduct glycomics studies on large populations6-9. This review introduces the new possibilities and expectations that large-scale glycome data obtained from cohort studies of healthy and diseased individuals may bring to glycoscience and the life sciences more broadly.
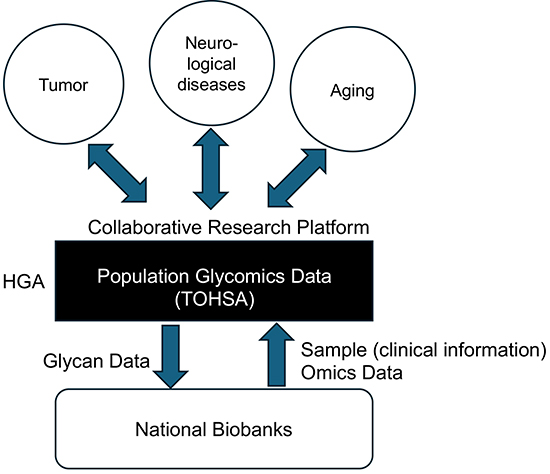
The Human Glycome Atlas Project (HGA) is a Japan-originated research initiative aimed at comprehensively mapping the human glycome and deepening our understanding of diseases and physiological processes involving glycans10,11. A unique and unprecedented aspect of the project is its focus on large-scale cohort-based glycomics studies. This initiative is supported by technological foundations that enable comprehensive analysis of the structurally diverse glycans, which add a new dimension to molecular diversity. In addition to integrated glycome analyses6,7,9, advanced methods such as robotics-assisted glycoproteomics8 and quantitative analysis of difficult-to-identify glycans have been introduced. In this article, we define this integrated and large-scale glycomics approach as “population glycomics” and review its outline and future prospects. We also introduce HGA's role as a collaborative research platform, highlighting its potential to foster interdisciplinary research across glycoscience and other scientific fields.
Glycans are widely recognized as disease-related molecules, and some specific glycan antigens have already been implemented as tumor markers12,13. Furthermore, the glycan structures attached to immunoglobulin G (IgG) have been shown to influence pathophysiology in infectious and autoimmune diseases14,15, indicating that glycans are not just biomarkers but also play functional roles in vivo. However, because glycans are added to many proteins, they represent an extremely diverse class of molecular modifications. Glycans exhibit a different type of diversity from genomes or proteins, introducing a new level of complexity to molecular heterogeneity. As a result, comprehensive glycan analysis remains technically challenging, and only a small portion of glycans have had their biological functions clarified. Against this backdrop, population glycomics in the HGA has the potential to greatly advance cohort-based studies linking glycans to various diseases. For example, by comparing glycomics results from disease groups to the glycan database provided by HGA, it becomes possible to identify biomarker candidates with differing abundance levels, enabling disease prediction. Moreover, early-phase glycan profiling may lead to earlier biomarker discovery and contribute to the development of novel interventions aimed at disease prevention. Indeed, early biomarker discovery is actively pursued in Alzheimer’s disease16, and glycomics may play a role in supporting the development of treatment strategies. The further development of interdisciplinary studies integrating the HGA glycan database is highly anticipated.
It has long been known that glycan expression varies even among healthy individuals, as exemplified by blood type differences. Large-scale analyses of IgG N-glycans across thousands of samples have revealed sex- and age-related differences in glycan expression17. Such variability likely extends beyond IgG to glycans in general, though comprehensive analyses on the same scale have not yet been conducted. HGA is planning large-scale glycome analyses using blood samples from thousands of healthy individuals in collaboration with Japanese biobanks, including the Tohoku Medical Megabank. These studies aim to characterize glycan profiles by sex and age, thereby enabling health assessments based on glycan information. Using these healthy profiles as standards will also support diverse applications. For example, by comparing pre-disease glycan profiles with clinical data, it will be possible to evaluate deviations from healthy baselines and predict disease risk based on glycan signatures. Such applications are expected to contribute to personalized medicine tailored to individual disease risks.
Large-scale glycome analyses within the HGA will reveal the overall glycan landscape in the Japanese population and enable the identification of disease biomarkers and risk indicators based on their associations with disease information. However, understanding the mechanisms by which glycans are involved in diseases requires integrated analysis with genomic data—the molecular foundation of disease—since glycan data alone are insufficient. Numerous genome-wide association studies (GWAS) have identified SNPs (single nucleotide polymorphisms) associated with many diseases and traits18. While GWAS targeting glycans on specific proteins—such as immunoglobulins—have been reported19, no comprehensive glycome GWAS analyses have been conducted to date. The HGA aims to promote glycan-focused GWAS by utilizing population glycomics data in conjunction with genomic information maintained by domestic biobanks, such as the National Center for Geriatrics and Gerontology and the Tohoku Medical Megabank Organization. Additionally, by applying epidemiological methods such as Mendelian randomization20, which uses SNPs commonly associated with both glycans and diseases as instrumental variables, it will become possible to explore causal relationships between glycans and diseases.
This article reviewed the concept and future potential of population glycomics as a core component of the large-scale cohort studies pursued by the HGA. The glycome profiles of large Japanese cohorts will be accumulated in a glycan database known as TOHSA and made widely accessible to the research community10. As a collaborative research platform, HGA is expected to catalyze progress across various research fields (see Figure 1). Furthermore, integration of glycan data with other omics data (genomics, proteomics, metabolomics, etc.) through collaboration with domestic biobanks will likely contribute to the development of more accurate and reliable disease biomarkers and health indicators.