
Hirokazu Yagi
Associate Professor, Graduate School of Pharmaceutical Sciences, Nagoya City University / Visiting Associate Professor, Exploratory Research Center on Life and Living Systems (ExCELLS), National Institutes of Natural Sciences
Hirokazu Yagi received his B.S. degree in Pharmaceutical Sciences from Nagoya City University in 2003 and completed his Ph.D. in 2008 at the Graduate School of Pharmaceutical Sciences, Nagoya City University. He subsequently served as a JSPS Research Fellow at both the same graduate school and the National Institute for Physiological Sciences. In 2009, he was appointed Assistant Professor at Nagoya City University, where he was later promoted to Lecturer and then Associate Professor. Since 2023, he has been leading his own research group as an independent Associate Professor. In addition, since 2022, he has concurrently held the position of Visiting Associate Professor at ExCELLS, National Institutes of Natural Sciences. His research focuses on elucidating the biosynthetic pathways and biological functions of glycans involved in various physiological processes.

Sachiko Kondo
Specially Appointed Senior Specialist, Exploratory Research Center on Life and Living Systems (ExCELLS), National Institutes of Natural Sciences
Sachiko Kondo received her M.Ed. in Science Education from the Graduate School of Education, Aichi University of Education in 2005. In the same year, she joined G LYENCE, a Nagoya City University-based biotech venture specializing in glycan structural analysis technologies. Since 2023, she has been serving in her current role at ExCELLS. Her work focuses on glycan structural analysis using HPLC mapping techniques.

Koichi Kato
Professor, Exploratory Research Center on Life and Living Systems (ExCELLS), National Institutes of Natural Sciences
Dr. Koichi Kato received his Ph.D. in Pharmaceutical Sciences from the University of Tokyo in 1991, after earning his bachelor's degree from the same university in 1986. He began his academic career at the University of Tokyo as an Assistant Professor and Lecturer. Since 2000, he has been a Professor at the Graduate School of Pharmaceutical Sciences, Nagoya City University. In 2008, he was appointed Professor at the Okazaki Institute for Integrative Bioscience, which was later reorganized into ExCELLS, where he has held his current position since 2018. He currently holds joint appointments at ExCELLS and Nagoya City University. His research focuses on elucidating the mechanisms of glycosylation through an integrated approach that spans molecular and cellular levels.
Glycan structural analysis using HPLC mapping enables the quantitative analysis of complex glycan structures while distinguishing isomers, playing an essential role in the Human Glycome Atlas Project. This article introduces the fundamental principles and applications of HPLC mapping, with a focus on its integration with mass spectrometry and the utilization of the "GALAXY" database for methods of identifying glycan structures. This technique significantly contributes to the acceleration of glycan research and the refinement of the human glycan map.
Glycans, due to their structural diversity, complexity, and inherent heterogeneity, present unique analytical challenges not encountered with other biological macromolecules. In high-throughput glycomics studies, including the Human Glycome Atlas Project, mass spectrometry (MS) has become the central technology for structural analysis. MS enables highly sensitive and accurate mass determination using minute sample amounts and, through tandem MS, allows rapid identification of monosaccharide compositions (e.g., Hex and HexNAc), their sequence, and glycosylation sites on proteins.
However, the biological functions of glycans often depend on subtle structural features such as linkages, branching patterns, stereochemistry, and terminal modifications, which are difficult to resolve by standard MS techniques. In particular, the separation and identification of isomers sharing the same monosaccharide composition remain major bottlenecks in glycan structure analysis.
In this context, glycan analysis using high-performance liquid chromatography (HPLC) developed by Takahashi and coworkers has played a crucial role. Based on differences in elution times, HPLC can discriminate structural isomers, making it highly effective for structural estimation and comparative analyses. The “HPLC map,” leveraging these properties, serves as a powerful tool for glycan structural identification1-4. Furthermore, when combined with MS and enzymatic treatments, HPLC mapping allows for the precise determination of chemical structure. This review outlines the principles and applications of glycan analysis using HPLC mapping.
In HPLC mapping, glycans are fluorescently labeled at their reducing ends using reagents such as 2-aminopyridine (PA), enabling highly sensitive detection. The labeled glycans are separated by HPLC, and their elution times are compared with known standards to identify structures (Figure 1). The retention time in HPLC is strongly dependent on glycan structure and is influenced by factors such as branching patterns, terminal modifications (e.g., fucosylation, galactosylation, and sialylation), phosphorylation or sulfation, and interactions with the fluorescent labels. These properties make HPLC mapping a powerful approach for visually and quantitatively capturing glycan structural information.
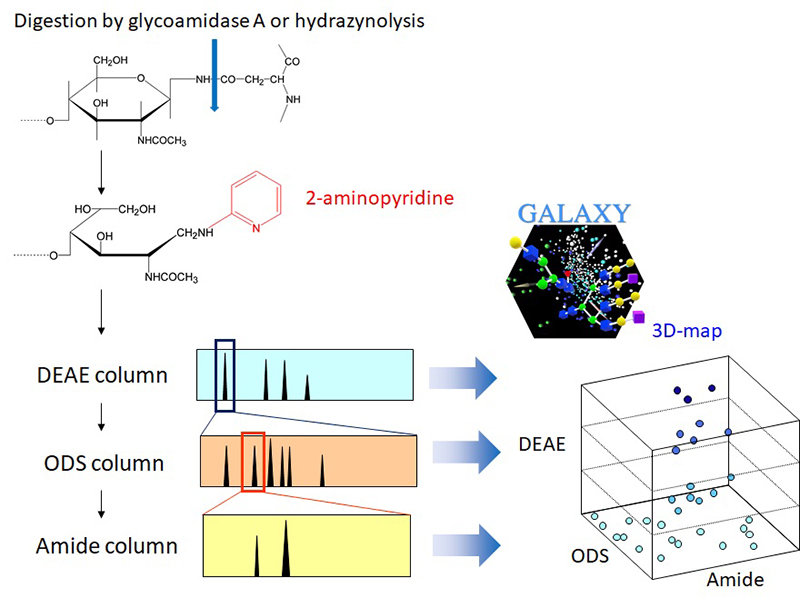
The three fundamental separation modes utilized in HPLC mapping are anion exchange chromatography, reversed-phase chromatography, and normal-phase chromatographies. In anion exchange chromatography, typically using diethylaminoethyl (DEAE) columns, acidic glycans containing sialic acid, sulfate, or glucuronic acid are separated according to their net negative charges.
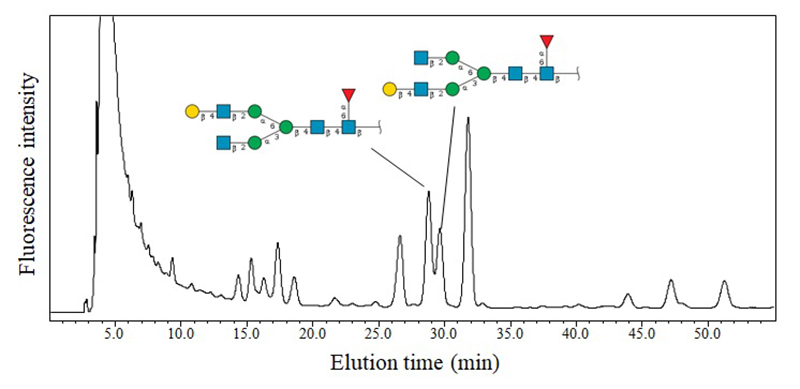
Reversed-phase chromatography, which employs octadecyl silica (ODS) columns, separates glycans based on subtle differences in hydrophobicity through interactions with hydrophobic packing materials. Optimization of elution conditions allows for the resolution and quantification of structural isomers that are otherwise difficult to distinguish by MS alone. For instance, Figure 2 shows a serum-derived N-glycan profile where isomers differing in galactose linkage positions are successfully separated and analyzed.
Normal-phase chromatography, utilizing amide columns, separates glycans according to hydrophilicity. In this mode, glycans with larger molecular weights and a higher number of hydroxyl groups tend to have longer retention times, resulting in a strong correlation between retention time and mass. By combining normal-phase HPLC with MS analysis, it becomes possible to further improve the throughput of glycan mapping.
To allow direct comparison between HPLC runs, retention times are standardized using glucose unit (GU) values, which are calculated based on the retention times of a standard glycan mixture. The use of GU values enables consistent comparisons across different days, instruments, and research facilities.
Moreover, HPLC mapping is not limited to the identification of known glycan structures. It also facilitates the exploration of unknown glycans by analyzing differences in retention time or the behavior of glycans after enzymatic treatments, providing valuable insights into their structural features. When combined with MS data, the precision of structural determination is greatly enhanced.
Because HPLC mapping uses fluorescence detection, it offers superior quantitative capability compared to MS and enables direct fractionation and collection of glycans after separation. These isolated glycans can then be used to construct glycan libraries, which are valuable resources for functional glycomics studies5.
Currently, a database containing HPLC data for over 600 known glycans has been established and is available through the web application "GALAXY"6,7. In GALAXY, users can input experimentally measured retention times and mass values to obtain a panel of candidate structures. Additionally, the platform predicts the likely elution positions of unknown glycans based on unit contribution values.
Since GALAXY is linked to the GlyCosmos Portal8, it allows seamless access to external databases, providing further information such as the three-dimensional structures of glycans and related glycoprotein data.
In high-throughput MS-based analyses, while monosaccharide compositions and sequences can be determined, it is generally difficult to definitively assign chemical structures. For N-glycans, approximate structures can be inferred based on biosynthetic knowledge, but strict identification of differences in linkage types—such as α2,3 versus α2,6 sialyl linkages, or β1,3 versus β1,4 galactosyl linkages—is challenging. Techniques such as SALSA (Sialic Acid Linkage-Specific Alkylamidation) exploit reactivity differences to distinguish sialyl linkages9, and multistage MSn analysis can also address isomer identification10. However, these approaches become complex and low-throughput when dealing with mixtures of isomers.
In contrast, HPLC mapping serves as a powerful complementary method to MS by facilitating the separation and identification of glycan isomers.
Moreover, the integration of MS improves both the accuracy and reliability of HPLC-based analyses. For example, by replacing elution time data from an amide column with mass information, it is possible to improve the accuracy of structural isomer identification while maintaining throughput. Similar to widely used LC-MS systems, coupling a mass spectrometer to the outlet of the ODS column employed in HPLC mapping allows for the collection of mass and structural information alongside retention time data for each glycan peak.
In the Human Glycome Atlas Project, an integrated approach combining MS composition data with HPLC-based chemical structure information has been adopted (Figure 3). By performing LC-MS analysis on glycopeptides containing individual glycosylation sites, the monosaccharide compositions associated with each site can be determined. Subsequently, applying HPLC mapping to glycopeptides separated by glycosylation site enables precise characterization of site-specific glycan structures, including information on isomerism.
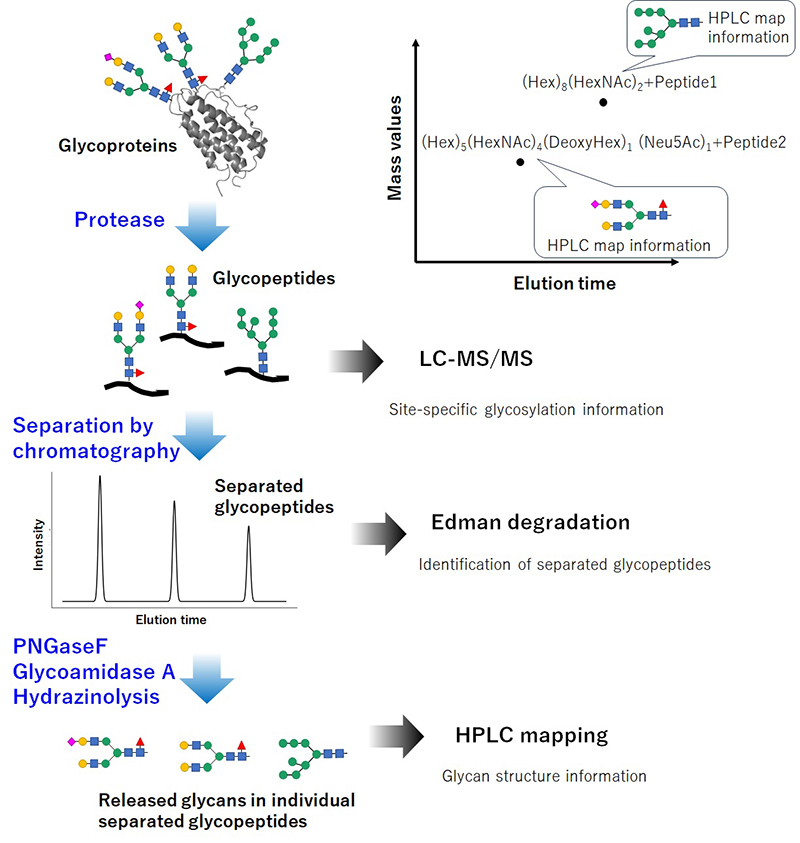
Glycan analysis using HPLC mapping is widely employed in glycoscience due to its high sensitivity, high resolution, quantitative capability, and visual/interpretive advantages. This method has been successfully applied in the identification of unknown glycan structures, quantitative comparisons, quality control of biopharmaceuticals, and biomarker discovery.
In the Human Glycome Atlas Project, HPLC mapping has played a critical role as a complementary technique to MS.
With ongoing expansion of HPLC-based glycan databases and the potential automation of sample preparation and analysis workflows, it is anticipated that large-scale glycan data accumulation will be further accelerated. Applying machine learning to these vast datasets could greatly enhance glycan structure identification and prediction. These outcomes not only contribute to the refinement of the human glycan map but also promote the comprehensive structural analysis and library creation of glycans found in nature, with the potential to further expand applications to the structural analysis of neo-glycans.